Virulence factors are molecules produced by pathogens that enhance their ability to infect and cause disease in host plants by overcoming plant defenses. In contrast, avirulence genes in pathogens encode proteins recognized by specific plant resistance genes, triggering defense responses that inhibit infection. The dynamic interaction between virulence factors and avirulence genes determines the outcome of host-pathogen interactions, influencing disease susceptibility or resistance.
Table of Comparison
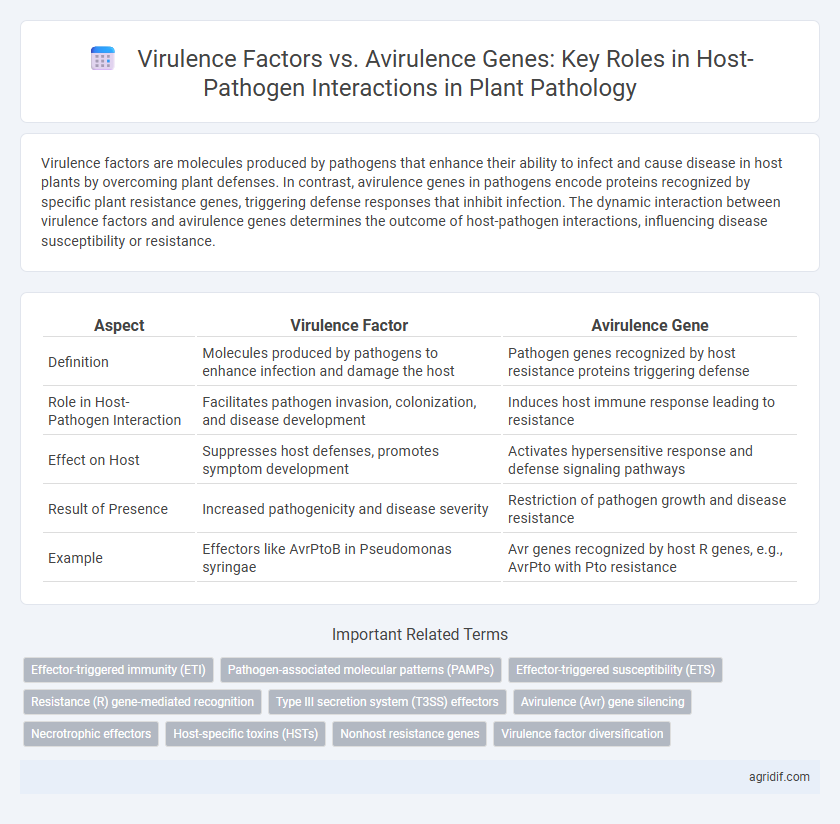
| Aspect | Virulence Factor | Avirulence Gene |
|---|---|---|
| Definition | Molecules produced by pathogens to enhance infection and damage the host | Pathogen genes recognized by host resistance proteins triggering defense |
| Role in Host-Pathogen Interaction | Facilitates pathogen invasion, colonization, and disease development | Induces host immune response leading to resistance |
| Effect on Host | Suppresses host defenses, promotes symptom development | Activates hypersensitive response and defense signaling pathways |
| Result of Presence | Increased pathogenicity and disease severity | Restriction of pathogen growth and disease resistance |
| Example | Effectors like AvrPtoB in Pseudomonas syringae | Avr genes recognized by host R genes, e.g., AvrPto with Pto resistance |
Defining Virulence Factors and Avirulence Genes
Virulence factors are molecules produced by pathogens that enable infection, colonization, and damage to the host plant by overcoming its immune defenses. Avirulence genes encode pathogen effectors recognized by specific plant resistance genes, triggering defense responses that limit pathogen success. These genetic components define the dynamic interaction between pathogen aggressiveness and host resistance in plant pathology.
Molecular Basis of Host-Pathogen Interactions
Virulence factors in pathogens encode proteins that suppress host immune responses and facilitate infection by targeting specific plant cellular mechanisms. In contrast, avirulence (Avr) genes trigger host resistance through recognition by plant resistance (R) proteins, activating defense pathways at a molecular level. The dynamic interplay between pathogen Avr genes and host R genes underlies gene-for-gene resistance, constituting a molecular basis for specificity in host-pathogen interactions.
Role of Virulence Factors in Pathogen Infection
Virulence factors are molecules produced by pathogens that facilitate colonization, suppress host immune responses, and promote nutrient acquisition, enabling successful infection and disease development in plants. These factors include enzymes, toxins, and effector proteins that disrupt host cellular processes and weaken plant defenses. Understanding the specific virulence factors involved in pathogen infection is crucial for developing resistant crop varieties and effective disease management strategies.
Function of Avirulence Genes in Plant Immunity
Avirulence genes in plant pathogens encode specific proteins recognized by corresponding resistance (R) genes in host plants, triggering effector-triggered immunity (ETI) that leads to localized cell death and restricts pathogen spread. These gene products act as signals, allowing plants to mount a defense response by activating hypersensitive response and systemic acquired resistance pathways. In contrast to virulence factors that promote infection, avirulence genes are key determinants in the genetic specificity of host-pathogen interactions, defining compatibility and resistance outcomes.
Gene-for-Gene Concept in Plant Pathology
Virulence factors are molecules produced by pathogens that enable infection and suppress host defenses, playing a critical role in overcoming plant immunity. Avirulence genes in pathogens encode specific effectors recognized by corresponding resistance (R) genes in the host, triggering defense mechanisms under the gene-for-gene concept. This interaction determines the outcome of host-pathogen compatibility, where the presence of matching avirulence genes and plant R genes leads to resistance, while their absence results in susceptibility.
Effector-Triggered Immunity vs Effector-Triggered Susceptibility
Virulence factors secreted by pathogens facilitate infection by suppressing the host's basal defenses, leading to effector-triggered susceptibility (ETS). In contrast, avirulence (Avr) genes encode effector proteins recognized by specific plant resistance (R) genes, activating effector-triggered immunity (ETI) that restricts pathogen growth. The dynamic interplay between pathogen effectors and host R proteins determines the outcome of host-pathogen interactions, influencing disease resistance or susceptibility.
Evolutionary Arms Race: Pathogen Adaptation and Plant Resistance
Virulence factors are molecules produced by pathogens that facilitate host invasion and suppress plant immune responses, enabling successful infection. Avirulence genes in pathogens encode effectors recognized by specific plant resistance (R) genes, triggering immune responses and preventing disease. The evolutionary arms race between pathogen virulence factors and plant avirulence genes drives coevolution, promoting pathogen adaptation and dynamic plant resistance mechanisms.
Mechanisms of Virulence Suppression by Avirulence Genes
Avirulence genes encode proteins that are recognized by specific host resistance (R) genes, triggering immune responses that suppress pathogen virulence factors. These avirulence gene products interfere with the pathogen's ability to deploy effectors that promote infection, effectively neutralizing virulence mechanisms such as effector-triggered susceptibility. The molecular recognition of avirulence gene effectors by host resistance proteins leads to hypersensitive response and systemic acquired resistance, limiting disease progression in plant-pathogen interactions.
Implications for Disease Resistance Breeding
Virulence factors in pathogens enable infection by overcoming host defenses, while avirulence genes trigger plant immune responses by being recognized by specific resistance (R) genes. Understanding the molecular interactions between virulence factors and avirulence genes informs targeted breeding strategies for durable disease resistance. Incorporating R genes corresponding to known avirulence genes enhances the effectiveness of resistance breeding programs against evolving pathogen populations.
Future Perspectives in Managing Plant Pathogens
Virulence factors enable pathogens to infect host plants by suppressing plant immune responses, while avirulence genes trigger host resistance through recognition mechanisms. Advances in genome editing and molecular breeding are poised to enhance the precision in manipulating these genetic components to develop durable disease-resistant crops. Integrating high-throughput sequencing and functional genomics will accelerate the identification of novel virulence and avirulence determinants, facilitating innovative strategies for sustainable plant pathogen management.
Related Important Terms
Effector-triggered immunity (ETI)
Virulence factors are molecules produced by pathogens that facilitate infection and suppress host defenses, while avirulence genes encode effectors recognized by specific plant resistance proteins, triggering Effector-triggered immunity (ETI). ETI activates robust defense responses in plants, often resulting in hypersensitive cell death to confine the pathogen and prevent disease progression.
Pathogen-associated molecular patterns (PAMPs)
Virulence factors enable pathogens to suppress host immune responses by interfering with the recognition of pathogen-associated molecular patterns (PAMPs), facilitating infection and colonization. Avirulence genes encode specific proteins recognized by host resistance genes, triggering effector-triggered immunity that counters PAMP-triggered immunity in plant-pathogen interactions.
Effector-triggered susceptibility (ETS)
Virulence factors are molecules produced by pathogens that suppress host immune responses, facilitating Effector-triggered susceptibility (ETS) by counteracting plant defense mechanisms, while avirulence genes encode effectors recognized by plant resistance proteins to trigger Effector-triggered immunity (ETI). The balance between virulence factor activity and avirulence gene recognition determines the outcome of host-pathogen interactions and susceptibility in plant pathology.
Resistance (R) gene-mediated recognition
Virulence factors are molecules produced by pathogens that facilitate infection and suppress host defense mechanisms, while avirulence (Avr) genes encode proteins recognized by specific resistance (R) genes in the host, triggering a defense response known as effector-triggered immunity (ETI). The interaction between Avr gene products and R gene-encoded receptors enables plants to detect pathogen presence and activate localized cell death to restrict pathogen spread.
Type III secretion system (T3SS) effectors
Virulence factors are effector proteins delivered by the Type III secretion system (T3SS) into host cells to suppress immune responses and promote pathogen colonization, whereas avirulence (Avr) genes encode effectors recognized by specific plant resistance proteins, triggering defense mechanisms. The interplay between T3SS effectors from virulence factors and host-encoded resistance genes defines the specificity of host-pathogen interactions and disease outcomes in plant pathology.
Avirulence (Avr) gene silencing
Avirulence (Avr) gene silencing in plant pathogens disables the recognition of specific host resistance (R) genes, allowing pathogens to evade immune responses and enhance virulence. This epigenetic regulation of Avr genes disrupts effector-triggered immunity, facilitating successful host colonization and disease progression.
Necrotrophic effectors
Necrotrophic effectors function as virulence factors by promoting host tissue necrosis to facilitate pathogen colonization, contrasting with avirulence genes that trigger host immune responses leading to resistance. These effectors manipulate host cell death pathways, enhancing pathogen aggressiveness in necrotrophic interactions.
Host-specific toxins (HSTs)
Virulence factors such as Host-Specific Toxins (HSTs) enable pathogens to infect and cause disease in specific host plants by targeting unique host cellular components, while avirulence genes in the pathogen encode effectors that can be recognized by host resistance genes, triggering defense responses. The interaction between HSTs and avirulence gene products determines the outcome of host-pathogen compatibility, influencing susceptibility or resistance in plant species.
Nonhost resistance genes
Virulence factors enable pathogens to infect specific hosts by overcoming host defenses, while avirulence genes in pathogens are recognized by nonhost resistance genes in plants, triggering immune responses that prevent infection. Nonhost resistance genes provide broad-spectrum, durable defense by detecting conserved avirulence effectors and activating resistance pathways, limiting pathogen adaptation.
Virulence factor diversification
Virulence factors evolve rapidly through gene duplication, point mutations, and horizontal gene transfer, enabling pathogens to overcome host defenses by targeting specific cellular processes. In contrast, avirulence genes trigger host immune responses when recognized, creating a selective pressure that drives continuous diversification of virulence factors to evade detection.
Virulence factor vs Avirulence gene for host-pathogen interaction Infographic
 agridif.com
agridif.com