Marker-assisted selection (MAS) leverages specific genetic markers linked to desirable traits to accelerate plant breeding, providing precise and cost-effective improvements in early-generation selection. Genomic selection (GS) utilizes genome-wide markers to predict breeding values, enabling more accurate and comprehensive advancement of complex traits influenced by multiple genes. Both methods enhance breeding efficiency, but GS offers greater predictive power for polygenic traits, while MAS remains valuable for traits controlled by major genes.
Table of Comparison
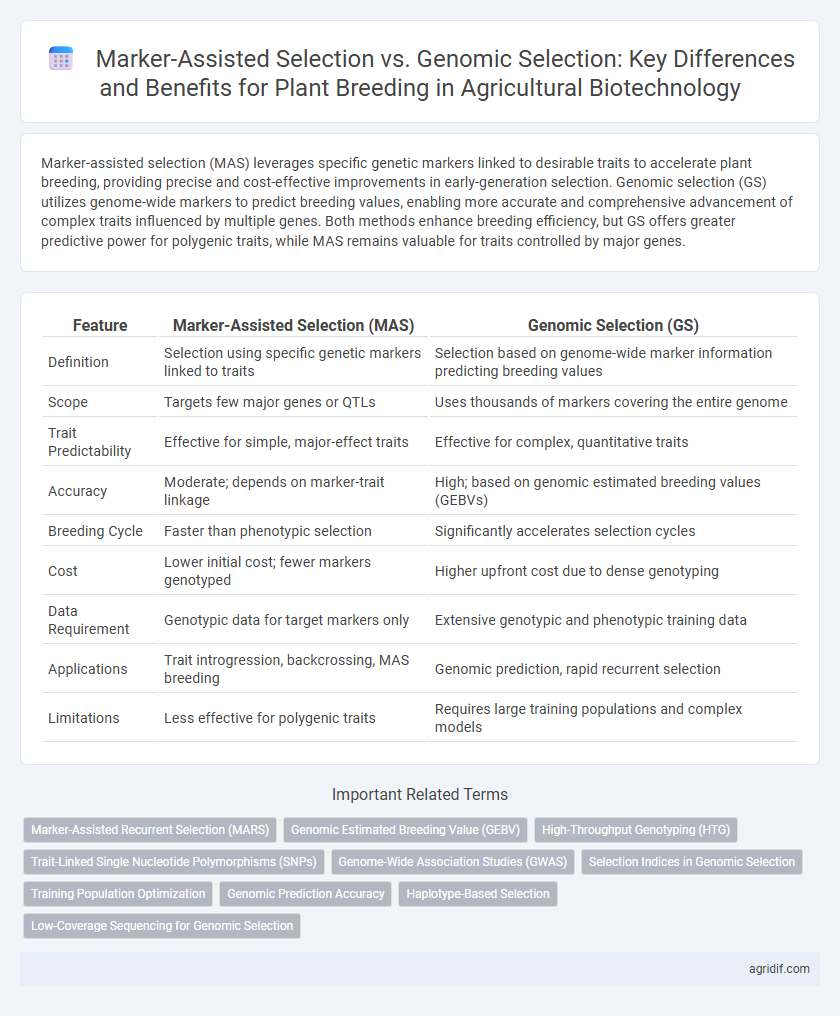
| Feature | Marker-Assisted Selection (MAS) | Genomic Selection (GS) |
|---|---|---|
| Definition | Selection using specific genetic markers linked to traits | Selection based on genome-wide marker information predicting breeding values |
| Scope | Targets few major genes or QTLs | Uses thousands of markers covering the entire genome |
| Trait Predictability | Effective for simple, major-effect traits | Effective for complex, quantitative traits |
| Accuracy | Moderate; depends on marker-trait linkage | High; based on genomic estimated breeding values (GEBVs) |
| Breeding Cycle | Faster than phenotypic selection | Significantly accelerates selection cycles |
| Cost | Lower initial cost; fewer markers genotyped | Higher upfront cost due to dense genotyping |
| Data Requirement | Genotypic data for target markers only | Extensive genotypic and phenotypic training data |
| Applications | Trait introgression, backcrossing, MAS breeding | Genomic prediction, rapid recurrent selection |
| Limitations | Less effective for polygenic traits | Requires large training populations and complex models |
Introduction to Modern Plant Breeding Technologies
Marker-assisted selection (MAS) utilizes specific DNA markers linked to desirable traits, enabling precise selection in early generations and reducing breeding cycles. Genomic selection (GS) employs genome-wide markers to predict breeding values, enhancing accuracy for complex traits with polygenic inheritance. Both technologies accelerate genetic gain and improve efficiency compared to traditional phenotypic selection in modern plant breeding programs.
Overview of Marker-Assisted Selection (MAS)
Marker-assisted selection (MAS) accelerates plant breeding by using molecular markers linked to desirable traits, enabling precise identification of plants carrying target genes without waiting for phenotypic expression. MAS improves efficiency in selecting traits such as disease resistance, yield, and stress tolerance by tracking specific genomic regions, reducing the breeding cycle time compared to traditional methods. While MAS targets a limited number of markers associated with major genes, it provides a cost-effective and accurate approach for incorporating known traits into new cultivars.
Fundamentals of Genomic Selection (GS)
Genomic Selection (GS) utilizes genome-wide marker data to predict the breeding values of plants, enabling more accurate and faster selection compared to Marker-Assisted Selection (MAS), which relies on a limited number of markers linked to specific traits. GS incorporates dense marker coverage to capture the effects of all quantitative trait loci (QTLs) simultaneously, improving prediction accuracy for complex traits like yield and stress tolerance. This approach accelerates genetic gain by enabling early selection in breeding cycles without phenotypic evaluation, optimizing resource use in plant breeding programs.
Key Differences Between MAS and GS
Marker-assisted selection (MAS) targets specific genetic markers linked to desirable traits, enabling breeders to select plants with known beneficial alleles efficiently. Genomic selection (GS) employs genome-wide markers to predict the breeding values of individual plants, capturing both major and minor gene effects for complex traits. MAS is ideal for traits controlled by a few genes, while GS excels in improving polygenic traits through comprehensive genetic evaluation.
Applications of MAS in Crop Improvement
Marker-assisted selection (MAS) accelerates crop improvement by enabling precise identification and selection of desirable traits linked to specific genetic markers, such as disease resistance or drought tolerance, in crops like wheat, maize, and rice. MAS facilitates introgression of beneficial alleles from wild relatives, enhancing yield stability and stress resilience in breeding programs. Applications of MAS in crop improvement include rapid pyramiding of multiple genes for biotic and abiotic stress resistance, improving nutrient use efficiency, and developing varieties with enhanced quality traits.
Role of GS in Accelerating Plant Breeding Programs
Genomic selection (GS) enhances plant breeding efficiency by using genome-wide markers to predict the genetic value of individuals, enabling early selection without phenotypic evaluation. Unlike marker-assisted selection (MAS), which relies on a few markers linked to specific traits, GS captures the effects of thousands of markers simultaneously, improving accuracy for complex traits. This approach significantly accelerates breeding cycles and increases genetic gain per unit of time in crops like maize and wheat.
Advantages and Limitations of Marker-Assisted Selection
Marker-assisted selection (MAS) enables targeted breeding by using specific DNA markers linked to desirable traits, accelerating the identification of plants with favorable genes and reducing breeding cycles. MAS is cost-effective for traits controlled by a few major genes but struggles with complex quantitative traits governed by multiple genes, limiting its accuracy compared to genomic selection. The primary limitation of MAS lies in its reliance on known markers and the difficulty in capturing the full genetic architecture of polygenic traits, whereas genomic selection offers a genome-wide approach that enhances prediction accuracy for complex traits.
Benefits and Challenges of Genomic Selection
Genomic selection accelerates plant breeding by using genome-wide markers to predict complex traits, enabling early and accurate selection of superior genotypes. This approach improves genetic gain per breeding cycle and enhances the efficiency of selecting traits controlled by multiple genes, such as yield and disease resistance. Challenges include the high cost of genotyping, the need for extensive training populations, and the complexity of data analysis requiring advanced bioinformatics tools.
Cost, Efficiency, and Scalability in MAS vs. GS
Marker-assisted selection (MAS) in plant breeding generally involves lower initial costs due to targeting specific, well-characterized genetic markers, but its efficiency is limited by the number of markers and traits analyzed. Genomic selection (GS) incurs higher upfront costs from genome-wide marker genotyping but achieves greater efficiency by capturing the effects of many small-effect loci, accelerating genetic gain per breeding cycle. Scalability favors GS as it leverages high-throughput genotyping platforms and statistical models to handle large populations and complex traits, whereas MAS can become labor-intensive and less scalable when multiple traits or loci are involved.
Future Prospects: Integrating MAS and GS for Sustainable Agriculture
Integrating marker-assisted selection (MAS) and genomic selection (GS) offers promising advancements for sustainable agriculture by enhancing genetic gain and breeding efficiency in crops. MAS targets specific genes linked to important traits, while GS evaluates entire genomes to predict performance, enabling the development of resilient and high-yield varieties under changing environmental conditions. Combining these approaches facilitates precise trait introgression and accelerates breeding cycles, supporting crop improvement for food security and climate adaptability.
Related Important Terms
Marker-Assisted Recurrent Selection (MARS)
Marker-Assisted Recurrent Selection (MARS) accelerates plant breeding by using molecular markers to track multiple desirable alleles across generations, enhancing genetic gain compared to traditional Marker-Assisted Selection (MAS). Genomic selection, however, utilizes genome-wide marker information and predictive models to capture complex trait variation, providing higher accuracy than MARS for polygenic traits in crop improvement.
Genomic Estimated Breeding Value (GEBV)
Genomic Estimated Breeding Value (GEBV) leverages genome-wide marker information to predict plant performance with higher accuracy compared to traditional Marker-Assisted Selection (MAS), which relies on a limited number of markers linked to specific traits. Utilizing GEBV in genomic selection accelerates breeding cycles and enhances selection efficiency by capturing the cumulative effects of thousands of single nucleotide polymorphisms (SNPs) across the entire genome.
High-Throughput Genotyping (HTG)
Marker-assisted selection (MAS) utilizes a limited number of genetic markers linked to specific traits, enabling targeted breeding decisions but often lacks genome-wide coverage, whereas genomic selection (GS) leverages High-Throughput Genotyping (HTG) technologies to analyze thousands of markers across the entire genome, improving prediction accuracy for complex traits in plant breeding. HTG platforms such as SNP arrays and next-generation sequencing facilitate the rapid and cost-effective genotyping of large breeding populations, making genomic selection a powerful strategy for accelerating genetic gain and enhancing crop improvement.
Trait-Linked Single Nucleotide Polymorphisms (SNPs)
Marker-assisted selection (MAS) leverages trait-linked single nucleotide polymorphisms (SNPs) to track specific gene variants associated with desired agronomic traits, enabling precise introgression in plant breeding. Genomic selection (GS) utilizes genome-wide SNP data to predict breeding values, capturing both major and minor effect loci, thereby accelerating selection for complex traits beyond the limitations of trait-specific markers.
Genome-Wide Association Studies (GWAS)
Marker-assisted selection (MAS) leverages Genome-Wide Association Studies (GWAS) to identify specific genetic markers linked to desirable plant traits, enabling targeted breeding with precision. Genomic selection extends beyond MAS by using genome-wide marker data from GWAS to predict the breeding value of plants, enhancing selection accuracy and accelerating genetic gain in crop improvement programs.
Selection Indices in Genomic Selection
Selection indices in genomic selection integrate multiple genomic estimated breeding values (GEBVs) weighted by their economic importance, optimizing the selection process for complex traits in plant breeding. This approach enhances genetic gain by enabling simultaneous improvement of yield, disease resistance, and stress tolerance, surpassing traditional marker-assisted selection's focus on a limited number of traits.
Training Population Optimization
Marker-assisted selection relies on a limited set of markers linked to key traits, making training population optimization focused on capturing genetic diversity around those loci, whereas genomic selection utilizes genome-wide markers, requiring a larger and more representative training population to enhance prediction accuracy across the entire genome. Effective training population optimization in genomic selection involves strategic sampling methods such as stratified or cluster sampling to maximize genetic variance and improve the reliability of genomic estimated breeding values.
Genomic Prediction Accuracy
Genomic selection outperforms marker-assisted selection by utilizing genome-wide marker data to predict breeding values with higher accuracy, capturing both major and minor gene effects. Enhanced genomic prediction accuracy accelerates genetic gain in plant breeding programs by enabling more precise selection decisions across diverse environments.
Haplotype-Based Selection
Haplotype-based selection in agricultural biotechnology leverages combinations of alleles at adjacent loci to improve accuracy in genomic selection compared to traditional marker-assisted selection, enabling more precise identification of beneficial traits in plant breeding. This method captures linkage disequilibrium patterns across haplotypes, enhancing prediction of complex traits such as yield and disease resistance, thereby accelerating genetic gain in crop improvement programs.
Low-Coverage Sequencing for Genomic Selection
Low-coverage sequencing in genomic selection enables cost-effective genotyping by capturing a broad spectrum of genome-wide marker information, increasing prediction accuracy for complex traits in plant breeding. Marker-assisted selection targets specific loci with known effects, but genomic selection with low-coverage data leverages genome-wide marker effects, enhancing genetic gain and selection efficiency in crop improvement.
Marker-assisted selection vs genomic selection for plant breeding Infographic
 agridif.com
agridif.com