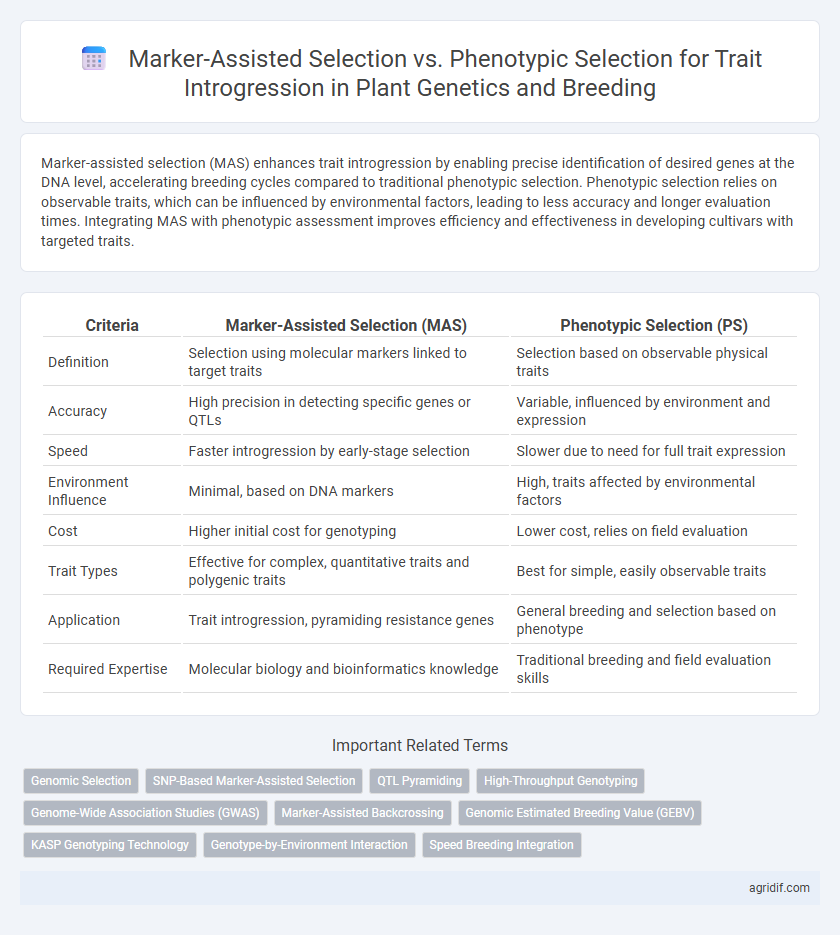
Marker-assisted selection (MAS) enhances trait introgression by enabling precise identification of desired genes at the DNA level, accelerating breeding cycles compared to traditional phenotypic selection. Phenotypic selection relies on observable traits, which can be influenced by environmental factors, leading to less accuracy and longer evaluation times. Integrating MAS with phenotypic assessment improves efficiency and effectiveness in developing cultivars with targeted traits.
Table of Comparison
| Criteria | Marker-Assisted Selection (MAS) | Phenotypic Selection (PS) |
|---|---|---|
| Definition | Selection using molecular markers linked to target traits | Selection based on observable physical traits |
| Accuracy | High precision in detecting specific genes or QTLs | Variable, influenced by environment and expression |
| Speed | Faster introgression by early-stage selection | Slower due to need for full trait expression |
| Environment Influence | Minimal, based on DNA markers | High, traits affected by environmental factors |
| Cost | Higher initial cost for genotyping | Lower cost, relies on field evaluation |
| Trait Types | Effective for complex, quantitative traits and polygenic traits | Best for simple, easily observable traits |
| Application | Trait introgression, pyramiding resistance genes | General breeding and selection based on phenotype |
| Required Expertise | Molecular biology and bioinformatics knowledge | Traditional breeding and field evaluation skills |
Introduction to Trait Introgression in Crop Improvement
Trait introgression involves transferring specific genes or quantitative trait loci (QTLs) from donor to recipient plant varieties to enhance crop performance. Marker-assisted selection (MAS) accelerates this process by using molecular markers tightly linked to target traits, enabling precise and efficient identification of desired genotypes without reliance on environmental conditions. Unlike phenotypic selection, which depends on observable traits that can be influenced by environmental variability, MAS improves the accuracy and speed of introgressing complex traits such as disease resistance and abiotic stress tolerance in crop improvement programs.
Fundamentals of Marker-Assisted Selection (MAS)
Marker-assisted selection (MAS) utilizes molecular markers linked to specific genes or quantitative trait loci (QTL) to accelerate the introgression of desired traits, enhancing precision over traditional phenotypic selection methods. MAS allows early and accurate identification of target traits at the seedling stage, reducing breeding cycles and environmental influence on trait expression. This genetic approach improves efficiency in selecting complex traits controlled by multiple genes, facilitating the development of superior crop varieties.
Principles of Phenotypic Selection in Plant Breeding
Phenotypic selection in plant breeding relies on observable traits to identify superior plants, emphasizing direct evaluation of phenotypic performance under specific environmental conditions. This method exploits genetic variation expressed through morphological, physiological, or biochemical characteristics to select individuals with desirable attributes for trait introgression. Phenotypic selection remains fundamental in diverse breeding programs despite advancements in marker-assisted selection, due to its simplicity and effectiveness in capturing complex trait interactions.
Genetic Basis and Efficiency of Trait Transfer
Marker-assisted selection (MAS) leverages specific genetic markers linked to desired traits, enabling precise and efficient introgression by directly targeting genomic regions controlling trait expression, while phenotypic selection relies on observable characteristics subject to environmental influence and polygenic effects. MAS enhances genetic gain by facilitating early and accurate selection, reducing linkage drag and accelerating trait incorporation in breeding populations. Phenotypic selection often requires multiple generations to achieve comparable precision, making MAS superior in terms of genetic resolution and efficiency for complex trait transfer.
Comparative Reliability: MAS vs. Phenotypic Selection
Marker-assisted selection (MAS) offers higher precision and reliability than phenotypic selection by directly targeting specific genes linked to desired traits, reducing the influence of environmental variability. Phenotypic selection relies on observable characteristics, which can be affected by external factors and may lead to less consistent trait introgression. MAS enables earlier and more accurate identification of plants carrying the target genes, accelerating breeding cycles and improving genetic gain efficiency.
Speed and Precision in Trait Introgression
Marker-assisted selection accelerates trait introgression by identifying desired alleles at the seedling stage, reducing the breeding cycle duration compared to phenotypic selection which relies on mature plant evaluation. This molecular approach enhances precision by ensuring the accurate incorporation of target genes while minimizing linkage drag, whereas phenotypic selection can be confounded by environmental variability and genotype-by-environment interactions. Consequently, marker-assisted selection provides faster and more reliable integration of complex traits in plant breeding programs.
Resource Use and Cost-effectiveness
Marker-assisted selection (MAS) enhances resource efficiency by enabling early and precise identification of desirable genotypes, significantly reducing time and labor compared to phenotypic selection. MAS minimizes the need for extensive field trials and large population screening, thereby lowering overall costs associated with trait introgression. Phenotypic selection often demands prolonged evaluation under variable environmental conditions, leading to higher resource consumption and increased expenses in breeding programs.
Challenges and Limitations of Each Approach
Marker-assisted selection faces challenges such as the high cost of genotyping and the complexity of identifying reliable markers for polygenic traits, limiting its effectiveness in some breeding programs. Phenotypic selection is constrained by environmental variability and the time-consuming nature of field evaluations, which can reduce selection accuracy and delay breeding cycles. Both approaches may struggle with linkage drag and background genome recovery, presenting significant hurdles in trait introgression efforts.
Case Studies: Successes in MAS and Phenotypic Selection
Marker-assisted selection (MAS) enhances trait introgression by precisely identifying and selecting target genes using molecular markers, as demonstrated in rice blast resistance breeding where MAS accelerated the introgression of resistance genes compared to phenotypic selection alone. Phenotypic selection remains effective in complex traits influenced by multiple genes and environmental factors, evidenced by drought-tolerant maize development through successive field evaluations and selection over multiple generations. Combining MAS with phenotypic selection in wheat quality improvement programs has yielded superior varieties by efficiently combining genomic data with observable trait performance.
Future Perspectives and Integration of Selection Methods
Marker-assisted selection (MAS) enhances precision in trait introgression by identifying specific genetic markers linked to desirable traits, accelerating breeding cycles compared to traditional phenotypic selection. Future perspectives emphasize integrating MAS with phenotypic evaluations to exploit both genotype and phenotype data, optimizing selection accuracy and resource efficiency. Combining these methods supports the development of superior cultivars with increased adaptability and yield stability under diverse environmental conditions.
Related Important Terms
Genomic Selection
Genomic selection accelerates trait introgression by utilizing genome-wide marker data to predict breeding values more accurately than traditional marker-assisted or phenotypic selection methods. This approach enhances genetic gain and selection efficiency by capturing small-effect loci across the entire genome, enabling rapid improvement of complex traits in plant breeding programs.
SNP-Based Marker-Assisted Selection
SNP-based marker-assisted selection (MAS) offers precise identification of desirable alleles, accelerating trait introgression compared to traditional phenotypic selection which relies on observable characteristics and is influenced by environmental factors. SNP markers enable high-throughput genotyping and early selection in breeding cycles, increasing efficiency and accuracy in incorporating complex traits such as disease resistance or drought tolerance.
QTL Pyramiding
Marker-assisted selection accelerates QTL pyramiding by precisely tracking multiple quantitative trait loci linked to desired traits, enhancing the efficiency of trait introgression compared to phenotypic selection which relies on observable characteristics and can be confounded by environmental factors. Integrating molecular markers enables simultaneous incorporation of diverse favorable alleles, improving genetic gain and reducing breeding cycles in plant breeding programs.
High-Throughput Genotyping
Marker-assisted selection leverages high-throughput genotyping technologies to rapidly and accurately identify desired alleles, significantly enhancing the efficiency of trait introgression compared to traditional phenotypic selection. This genotyping-driven approach enables precise selection at the DNA level, reducing breeding cycles and increasing the success rate of incorporating complex traits into plant genomes.
Genome-Wide Association Studies (GWAS)
Genome-wide association studies (GWAS) enable precise identification of trait-linked genetic markers, significantly enhancing the efficiency of marker-assisted selection (MAS) over traditional phenotypic selection for trait introgression in plant breeding. MAS accelerates the introgression process by targeting specific alleles responsible for desirable traits, reducing the need for extensive field evaluations inherent in phenotypic selection.
Marker-Assisted Backcrossing
Marker-assisted backcrossing (MAB) accelerates trait introgression by using molecular markers to precisely track target genes, reducing linkage drag and improving selection accuracy compared to traditional phenotypic selection. This genomic approach enhances breeding efficiency, enabling faster recovery of the recurrent parent's genome while ensuring the incorporation of desired traits.
Genomic Estimated Breeding Value (GEBV)
Marker-assisted selection (MAS) enhances trait introgression efficiency by using Genomic Estimated Breeding Value (GEBV) to predict genetic potential based on molecular markers, accelerating genetic gain compared to phenotypic selection. GEBV integrates genome-wide marker data, enabling precise identification of superior genotypes and reducing reliance on observable traits influenced by environmental variability.
KASP Genotyping Technology
Marker-assisted selection using KASP genotyping technology enables precise identification of desirable alleles at the seedling stage, significantly accelerating trait introgression compared to conventional phenotypic selection, which relies on observable traits that may be influenced by environmental factors. KASP assays offer high-throughput, cost-effective, and accurate genotyping, enhancing selection efficiency in breeding programs by targeting specific genetic markers linked to key agronomic traits.
Genotype-by-Environment Interaction
Marker-assisted selection (MAS) enables precise introgression of target genes by leveraging molecular markers, minimizing the confounding effects of genotype-by-environment interaction (GxE) that often obscure phenotypic selection outcomes. Unlike phenotypic selection, MAS enhances selection accuracy and efficiency across diverse environments by directly targeting genotypic variants linked to desirable traits, thereby accelerating genetic gain in plant breeding programs.
Speed Breeding Integration
Marker-assisted selection accelerates trait introgression by enabling precise identification of desired alleles at early seedling stages, significantly reducing breeding cycles when integrated with speed breeding protocols that shorten generation time to weeks. Phenotypic selection relies on observable traits, often limited by environmental variability and longer generational intervals, making it less efficient for rapid genetic gain compared to the synergy of marker-assisted selection and speed breeding techniques.
Marker-assisted selection vs phenotypic selection for trait introgression Infographic
 agridif.com
agridif.com