Narrow sense heritability measures the proportion of phenotypic variance attributed to additive genetic factors directly passed from parents to offspring, making it crucial for predicting response to selection in plant breeding. Broad sense heritability encompasses all genetic variance components, including additive, dominance, and epistatic interactions, providing insight into the total genetic influence on trait expression. Understanding the distinction between these two heritability concepts enables breeders to design effective selection strategies for improving complex traits in crops.
Table of Comparison
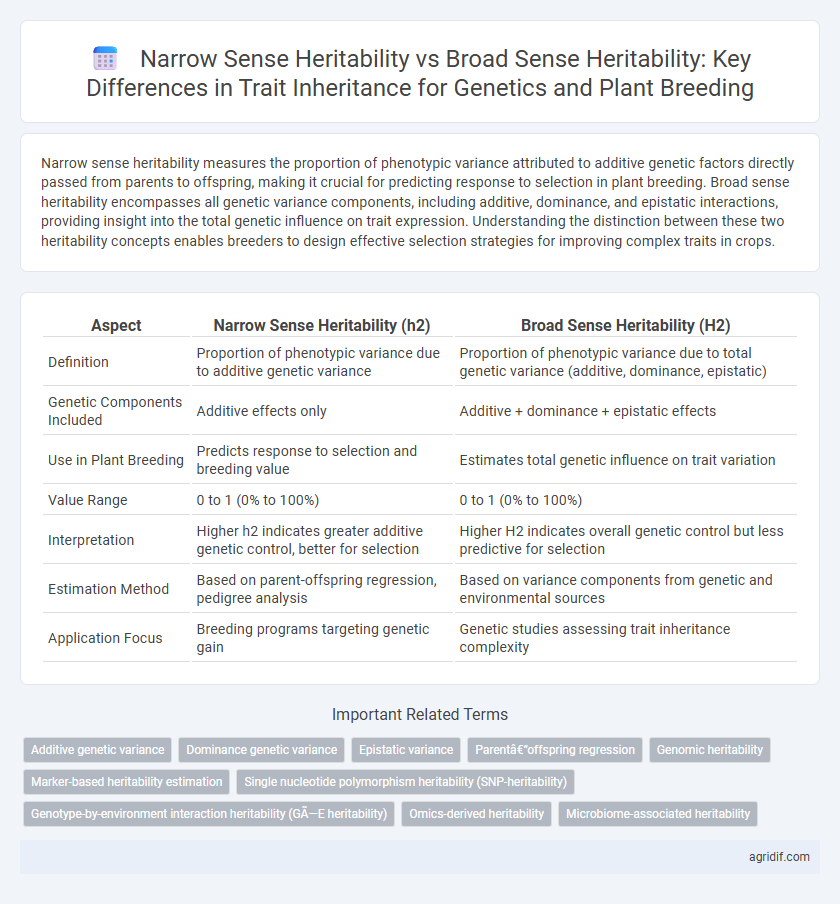
| Aspect | Narrow Sense Heritability (h2) | Broad Sense Heritability (H2) |
|---|---|---|
| Definition | Proportion of phenotypic variance due to additive genetic variance | Proportion of phenotypic variance due to total genetic variance (additive, dominance, epistatic) |
| Genetic Components Included | Additive effects only | Additive + dominance + epistatic effects |
| Use in Plant Breeding | Predicts response to selection and breeding value | Estimates total genetic influence on trait variation |
| Value Range | 0 to 1 (0% to 100%) | 0 to 1 (0% to 100%) |
| Interpretation | Higher h2 indicates greater additive genetic control, better for selection | Higher H2 indicates overall genetic control but less predictive for selection |
| Estimation Method | Based on parent-offspring regression, pedigree analysis | Based on variance components from genetic and environmental sources |
| Application Focus | Breeding programs targeting genetic gain | Genetic studies assessing trait inheritance complexity |
Introduction to Heritability in Plant Breeding
Narrow sense heritability measures the proportion of phenotypic variance attributed to additive genetic variance, crucial for predicting response to selection in plant breeding. Broad sense heritability encompasses all genetic variance components, including additive, dominance, and epistatic effects, providing an overall estimate of genetic influence on trait inheritance. Understanding both metrics aids breeders in optimizing selection strategies and improving trait transmission efficiency.
Defining Narrow Sense Heritability (h²)
Narrow sense heritability (h2) quantifies the proportion of phenotypic variance attributable to additive genetic variance, critical for predicting response to selection in plant breeding. It contrasts with broad sense heritability, which includes additive, dominance, and epistatic variance, but h2 specifically measures the transmissible genetic component from parents to offspring. Accurate estimation of narrow sense heritability enables breeders to optimize selection strategies for traits like yield, disease resistance, and stress tolerance.
Defining Broad Sense Heritability (H²)
Broad sense heritability (H2) quantifies the proportion of total phenotypic variance in a trait attributable to all genetic factors, including additive, dominance, and epistatic variances. It is calculated as the ratio of genetic variance (V_G) to the total phenotypic variance (V_P), expressed as H2 = V_G / V_P. This measure captures the overall genetic contribution to trait inheritance, informing breeding strategies that exploit both additive and non-additive genetic effects.
Genetic Variance Components: Additive, Dominance, and Epistasis
Narrow sense heritability (h2) quantifies the proportion of phenotypic variance attributable exclusively to additive genetic variance, essential for predicting response to selection in plant breeding. Broad sense heritability (H2) encompasses total genetic variance, including additive, dominance, and epistatic interactions, reflecting overall genetic influence on trait expression but less predictive for selection efficiency. Understanding the contributions of additive, dominance, and epistatic variance components enables breeders to optimize strategies for trait improvement, balancing genetic gain and maintaining genetic diversity.
Calculating Narrow Sense Heritability
Narrow sense heritability (h2) quantifies the proportion of phenotypic variance attributable to additive genetic variance, making it crucial for predicting response to selection in plant breeding. It is calculated by dividing the additive genetic variance (Va) by the total phenotypic variance (Vp), expressed as h2 = Va/Vp. This measure contrasts with broad sense heritability (H2), which includes all genetic variance components, thus providing a more precise estimate for the inheritance of traits influenced primarily by additive effects.
Calculating Broad Sense Heritability
Broad sense heritability (H2) quantifies the proportion of total phenotypic variance attributable to genetic variance, including additive, dominance, and epistatic effects. It is calculated using the formula H2 = VG / VP, where VG represents total genetic variance and VP represents total phenotypic variance, often estimated from variance components obtained through ANOVA in plant breeding experiments. Precise estimation of broad sense heritability aids in understanding the genetic contribution to trait inheritance, crucial for selecting breeding strategies.
Implications for Trait Selection and Improvement
Narrow sense heritability (h2) measures the proportion of phenotypic variance attributed to additive genetic variance, directly influencing the response to selection in quantitative traits. Broad sense heritability (H2) encompasses total genetic variance, including additive, dominance, and epistatic effects, offering insight into overall genetic contribution but not predicting selection efficiency as precisely. High narrow sense heritability facilitates effective selection and genetic gain in plant breeding, while reliance on broad sense heritability may overestimate heritable potential for trait improvement due to non-additive genetic components.
Limitations of Narrow and Broad Sense Heritability
Narrow sense heritability captures the proportion of phenotypic variance attributable to additive genetic variance, making it essential for predicting response to selection but it excludes dominance and epistatic effects, limiting its scope in complex trait inheritance. Broad sense heritability encompasses all genetic variance components--additive, dominance, and epistatic--but its overestimation of heritable variation can mislead breeding decisions because it does not separate these components. Both measures rely on assumptions such as no genotype-environment interaction and random mating, which are often violated in natural populations, reducing their predictive accuracy in practical plant breeding scenarios.
Heritability Estimates in Various Crop Traits
Narrow sense heritability (h2) measures the proportion of phenotypic variance attributed to additive genetic variance, crucial for predicting response to selection in plant breeding programs. Broad sense heritability (H2) encompasses all genetic variance components, including additive, dominance, and epistatic effects, providing insight into total genetic contribution to trait variation. Heritability estimates vary among crop traits, with narrow sense heritability often lower than broad sense due to the exclusion of non-additive effects, influencing breeding strategies for yield, disease resistance, and quality traits.
Practical Applications in Plant Breeding Programs
Narrow sense heritability (h2) estimates the proportion of phenotypic variance due to additive genetic factors, directly influencing selection response in plant breeding programs. Broad sense heritability (H2) encompasses total genetic variance, including additive, dominance, and epistatic effects, providing an overall measure of trait inheritance but less predictive power for selection gain. Practical breeding decisions rely heavily on narrow sense heritability to efficiently select superior genotypes and accelerate genetic improvement in crops.
Related Important Terms
Additive genetic variance
Narrow sense heritability measures the proportion of phenotypic variance attributable specifically to additive genetic variance, which directly influences the response to selection in plant breeding programs. Broad sense heritability encompasses all genetic variance components, including additive, dominance, and epistatic variances, providing a general estimate of trait inheritance but less predictive power for additive genetic gain.
Dominance genetic variance
Narrow sense heritability (h2) measures the proportion of phenotypic variance attributable to additive genetic variance, excluding dominance and epistatic effects, making it crucial for predicting response to selection in plant breeding. Broad sense heritability (H2) encompasses total genetic variance, including additive, dominance, and epistatic components, reflecting overall genetic influence on trait inheritance but less predictive for breeding progress where dominance genetic variance plays a significant role.
Epistatic variance
Narrow sense heritability measures the proportion of phenotypic variance attributed to additive genetic effects, excluding epistatic interactions, while broad sense heritability encompasses total genetic variance, including additive, dominance, and epistatic variance. Epistatic variance significantly influences broad sense heritability by capturing gene-gene interactions that contribute to complex trait inheritance beyond additive effects.
Parent–offspring regression
Narrow sense heritability (h2) quantifies the proportion of phenotypic variance attributable to additive genetic variance, making it central to predicting trait response through parent-offspring regression in plant breeding. Broad sense heritability (H2) encompasses total genetic variance, including additive, dominance, and epistatic effects, but parent-offspring regression primarily estimates narrow sense heritability for effective selection of inheritable traits.
Genomic heritability
Narrow sense heritability measures the proportion of phenotypic variance attributed to additive genetic variance, essential for predicting response to selection in plant breeding, while broad sense heritability encompasses total genetic variance, including dominance and epistatic effects. Genomic heritability, estimated using genome-wide markers, provides a more precise capture of additive genetic effects compared to traditional pedigree-based methods, enhancing the accuracy of trait inheritance predictions in quantitative genetics.
Marker-based heritability estimation
Narrow sense heritability quantifies the proportion of phenotypic variance attributable to additive genetic effects, crucial for predicting response to selection in plant breeding, while broad sense heritability encompasses all genetic variance including additive, dominance, and epistatic effects. Marker-based heritability estimation leverages molecular markers to more accurately partition additive genetic variance, improving precision in selecting traits with higher narrow sense heritability for genetic gain in breeding programs.
Single nucleotide polymorphism heritability (SNP-heritability)
Narrow sense heritability estimates the proportion of phenotypic variance attributed to additive genetic effects, essential for predicting response to selection in plant breeding, while broad sense heritability includes additive, dominance, and epistatic effects, reflecting total genetic variance. SNP-heritability quantifies the contribution of single nucleotide polymorphisms to narrow sense heritability, providing a precise genomic estimation of additive genetic variance for complex trait inheritance in plants.
Genotype-by-environment interaction heritability (G×E heritability)
Narrow sense heritability estimates the proportion of phenotypic variance attributable to additive genetic effects, crucial for predicting selection response in breeding programs, whereas broad sense heritability encompasses all genetic variance, including additive, dominance, and epistatic effects. Genotype-by-environment interaction heritability (GxE heritability) quantifies the genetic variance that arises from differential genotype performance across environments, significantly influencing trait stability and adaptive breeding strategies.
Omics-derived heritability
Narrow sense heritability, representing the proportion of phenotypic variance attributed to additive genetic effects, is crucial for predicting response to selection in plant breeding programs, whereas broad sense heritability encompasses additive, dominance, and epistatic variance, providing a comprehensive measure of genetic influence on a trait. Omics-derived heritability integrates genomics, transcriptomics, and epigenomics data, enhancing the precision of heritability estimates by capturing molecular-level variations that contribute to both narrow and broad sense heritability in trait inheritance.
Microbiome-associated heritability
Narrow sense heritability measures the proportion of phenotypic variance attributable to additive genetic factors influencing microbiome-associated traits, crucial for predicting response to selection in plant breeding. Broad sense heritability includes additive, dominance, and epistatic genetic variances, capturing the total genetic contribution to microbiome-driven trait inheritance, though it is less predictive for selection outcomes.
Narrow sense heritability vs Broad sense heritability for trait inheritance Infographic
 agridif.com
agridif.com