Oomycetes differ from true fungi primarily in their cell wall composition, with oomycetes having cellulose-based walls while true fungi possess chitin. Oomycetes are classified under the kingdom Stramenopila, whereas true fungi belong to the kingdom Fungi, reflecting fundamental genetic and biochemical differences. These distinctions impact pathogen classification and influence the strategies used for disease control in plant pathology.
Table of Comparison
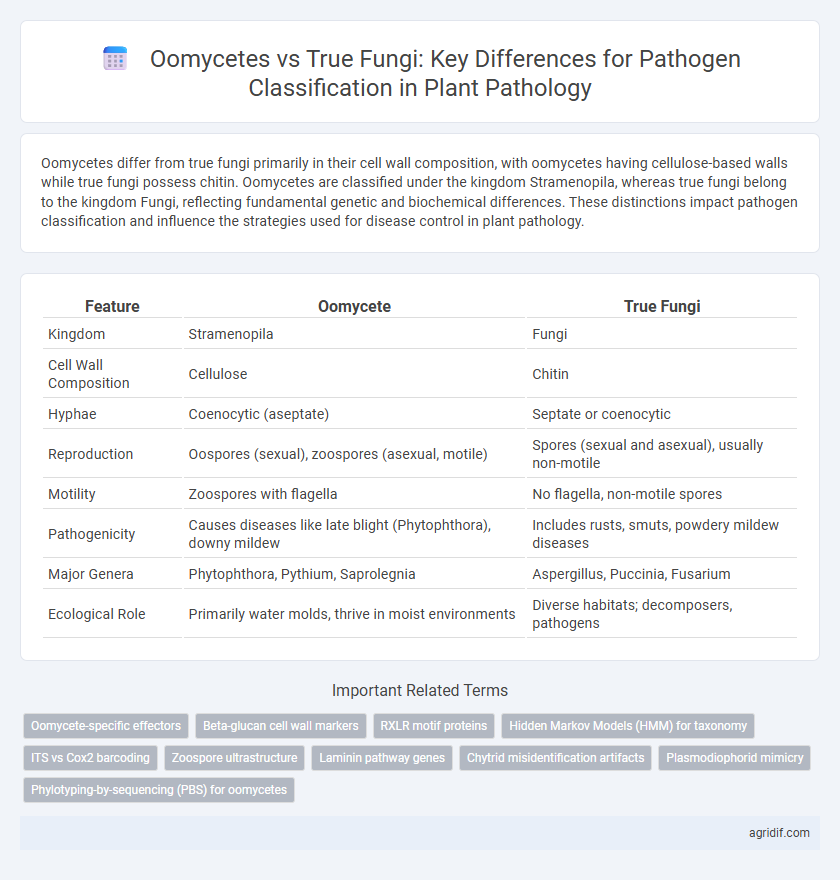
| Feature | Oomycete | True Fungi |
|---|---|---|
| Kingdom | Stramenopila | Fungi |
| Cell Wall Composition | Cellulose | Chitin |
| Hyphae | Coenocytic (aseptate) | Septate or coenocytic |
| Reproduction | Oospores (sexual), zoospores (asexual, motile) | Spores (sexual and asexual), usually non-motile |
| Motility | Zoospores with flagella | No flagella, non-motile spores |
| Pathogenicity | Causes diseases like late blight (Phytophthora), downy mildew | Includes rusts, smuts, powdery mildew diseases |
| Major Genera | Phytophthora, Pythium, Saprolegnia | Aspergillus, Puccinia, Fusarium |
| Ecological Role | Primarily water molds, thrive in moist environments | Diverse habitats; decomposers, pathogens |
Introduction to Oomycetes and True Fungi in Plant Pathology
Oomycetes and true fungi are distinct groups of pathogens crucial in plant pathology, with oomycetes belonging to the kingdom Stramenopila and true fungi classified under the kingdom Fungi. Oomycetes exhibit cell walls composed mainly of cellulose and produce biflagellate zoospores, whereas true fungi have chitin-based cell walls and reproduce through spores without flagella. The differentiation between oomycetes and true fungi is essential for accurate diagnosis and effective management of plant diseases caused by these pathogens.
Historical Perspectives on Pathogen Classification
Oomycetes, historically grouped with fungi due to morphological similarities, were later reclassified based on molecular phylogenetics distinguishing their cellulose-rich cell walls and diploid nuclei from the chitinous walls and haploid nuclei of true fungi. Early pathogen classification relied heavily on visible structures and reproduction methods, leading to misidentification between these two groups. Advances in genetic analysis have clarified their evolutionary divergence, significantly impacting disease management strategies in plant pathology.
Morphological Differences Between Oomycetes and True Fungi
Oomycetes, often called water molds, possess coenocytic hyphae lacking septa, whereas true fungi feature septate or aseptate hyphae depending on the species. The cell walls of oomycetes are primarily composed of cellulose and beta-glucans, contrasting with true fungi that have chitin-based cell walls. Oomycetes produce motile zoospores with two flagella, a characteristic absent in true fungi, which typically disperse via non-motile spores.
Cell Wall Composition: Oomycetes vs True Fungi
Oomycetes possess cell walls primarily composed of cellulose and beta-glucans, distinguishing them from true fungi, which have chitin-based cell walls. This fundamental difference in cell wall composition affects their structural integrity and susceptibility to antifungal agents. Understanding the cellulose-rich cell walls of oomycetes is crucial for developing targeted disease management strategies in plant pathology.
Reproductive Strategies and Life Cycles
Oomycetes differ from true fungi in reproductive strategies by producing biflagellate zoospores during asexual reproduction, facilitating waterborne dispersal, whereas true fungi primarily generate non-motile spores such as conidia or sporangia. In sexual reproduction, oomycetes form oospores through the fusion of distinct antheridia and oogonia, exhibiting a diploid dominant life cycle, contrasting sharply with the haploid-dominant cycles and formation of zygospores, ascospores, or basidiospores seen in true fungi. These differences in life cycle stages and spore morphology are critical for pathogen classification and understanding their epidemiology in plant diseases.
Molecular Phylogeny and Evolutionary Relationships
Oomycetes belong to the Stramenopiles group, exhibiting distinct molecular phylogenetic markers such as unique mitochondrial DNA sequences and cellulose-rich cell walls, contrasting with chitin-containing walls of true fungi in the Kingdom Fungi. Molecular phylogeny based on rRNA gene sequences and multi-locus analyses consistently separate Oomycetes from true fungi, reflecting their divergent evolutionary origins dating back to the divergence of Chromalveolata and Opisthokonta supergroups. Evolutionary relationships indicate Oomycetes adapted pathogenic lifestyles independently, resulting in convergent pathogenic traits despite distinct ancestral lineages compared to true fungal pathogens.
Disease Symptoms and Host Interactions
Oomycetes, unlike true fungi, often cause water-soaked lesions and rapidly spreading root rots due to their motile zoospores that infect host plants, leading to symptoms such as damping-off and downy mildew. True fungi generally form distinct mycelial growths producing visible spores, resulting in symptoms like powdery mildew, rusts, and cankers that develop more gradually. Host interactions with oomycetes typically involve aggressive tissue invasion facilitated by cell wall-degrading enzymes, whereas true fungi engage in a broader range of symbiotic and pathogenic relationships mediated through specialized infection structures like haustoria.
Diagnostic Techniques for Pathogen Identification
Oomycetes differ from true fungi in pathogen classification by their distinct cell wall composition and reproductive structures, necessitating specialized diagnostic techniques such as PCR-based assays targeting specific genetic markers like the ITS region. Immunofluorescence and monoclonal antibody tests enhance rapid detection of oomycete pathogens, while traditional fungal stains often fail due to differing cell wall polysaccharides. Molecular diagnostics combined with microscopic examination provide accurate differentiation critical for effective disease management in plant pathology.
Implications for Disease Management in Agriculture
Oomycetes, distinct from true fungi due to their cellulose-rich cell walls and diploid nuclei, require targeted fungicides like metalaxyl, as traditional fungicides effective against true fungi often fail. Accurate pathogen classification between oomycetes and true fungi informs the deployment of resistant crop varieties and the timing of irrigation to minimize disease spread. Integrating molecular diagnostics enhances early detection, enabling precise and sustainable disease management strategies in agriculture.
Future Directions in Oomycete and Fungal Pathogen Research
Future research in plant pathology emphasizes genomic and transcriptomic analyses to unravel distinct pathogenic mechanisms between oomycetes and true fungi, fostering targeted disease control strategies. Advances in CRISPR-Cas9 gene editing and RNA interference hold promise for functional studies of effector proteins unique to Phytophthora species and fungal pathogens like Fusarium oxysporum. Integration of multi-omics data and machine learning models will enhance prediction and management of host-pathogen interactions, ultimately improving crop resistance breeding programs.
Related Important Terms
Oomycete-specific effectors
Oomycetes, distinct from true fungi, possess unique effector proteins such as RXLR and CRN effectors that manipulate host plant immune responses to facilitate infection. These Oomycete-specific effectors are crucial molecular tools for pathogenicity, differentiating them from fungal pathogens which use different effector families like LysM effectors.
Beta-glucan cell wall markers
Oomycetes possess cell walls primarily composed of beta-glucans and cellulose, distinguishing them from true fungi, whose cell walls are predominantly chitin-based with beta-glucans as secondary components. This difference in beta-glucan composition serves as a critical molecular marker for pathogen classification in plant pathology, aiding in accurate diagnosis and targeted disease management strategies.
RXLR motif proteins
RXLR motif proteins are key effectors in Oomycete pathogens, facilitating host cell entry and immune system manipulation, whereas true fungi lack this conserved RXLR motif, relying on different effector proteins for pathogenicity. Understanding the distinct RXLR effector mechanisms aids in accurate pathogen classification and targeted disease management in plant pathology.
Hidden Markov Models (HMM) for taxonomy
Hidden Markov Models (HMM) enhance the taxonomy of plant pathogens by accurately distinguishing Oomycetes from true fungi based on unique molecular sequence patterns, leveraging their divergent evolutionary lineages within eukaryotes. Applying HMM to genomic and proteomic data reveals distinct motifs and domain architectures critical for classifying pathogenic species, improving diagnostic precision in plant disease management.
ITS vs Cox2 barcoding
ITS (Internal Transcribed Spacer) regions serve as the primary DNA barcode for classifying true fungi due to their high variability and species-level resolution, while Cox2 (cytochrome c oxidase subunit 2) gene sequences provide more reliable identification for oomycetes, a phylogenetically distinct group often misclassified as fungi. Comparative analyses reveal that ITS barcoding may yield ambiguous results in oomycetes, where Cox2 markers offer enhanced specificity and phylogenetic clarity in plant pathogen diagnostics.
Zoospore ultrastructure
Zoospore ultrastructure in oomycetes is characterized by the presence of a single posterior whiplash flagellum and a unique cytoskeletal arrangement with tubular mitochondrial cristae, distinguishing them from true fungi, which lack flagella and have lamellar mitochondrial cristae. This ultrastructural difference reflects their divergent evolutionary lineages, with oomycetes belonging to the Stramenopiles and true fungi classified under the Opisthokonts, impacting pathogen classification and disease management strategies.
Laminin pathway genes
Oomycetes, distinct from true fungi, possess unique laminin pathway genes that play critical roles in host-pathogen interactions and adhesion mechanisms, highlighting their divergent evolutionary lineage. These laminin-related genes contribute to the pathogen's ability to breach plant defenses, making them pivotal targets in plant pathology research for disease management strategies.
Chytrid misidentification artifacts
Oomycetes, often mistaken for true fungi due to similar filamentous growth and spore production, differ fundamentally in cell wall composition, primarily containing cellulose instead of chitin, which is characteristic of true fungi. Misidentification of chytrids arises from overlapping morphological traits, but molecular markers such as rRNA gene sequencing and zoospore ultrastructure enable accurate classification by distinguishing chytrids from oomycete pathogens.
Plasmodiophorid mimicry
Oomycetes, distinct from true fungi, possess cellulose in their cell walls and diploid-dominant life cycles, contrasting with the chitin-based walls and haploid dominance of true fungi, complicating pathogen classification in plant pathology. Plasmodiophorids, often mimicking fungi morphologically yet classified as obligate parasite protists within Cercozoa, further challenge traditional fungal pathogen identification by exhibiting unique endoparasitic traits in root tissues.
Phylotyping-by-sequencing (PBS) for oomycetes
Phylotyping-by-sequencing (PBS) enables precise differentiation between oomycetes and true fungi by targeting species-specific genetic markers unique to oomycetes, enhancing pathogen identification accuracy in plant pathology. This molecular approach overcomes limitations of traditional morphology-based classification by providing high-resolution phylogenetic data critical for managing oomycete-induced diseases.
Oomycete vs True Fungi for pathogen classification Infographic
 agridif.com
agridif.com