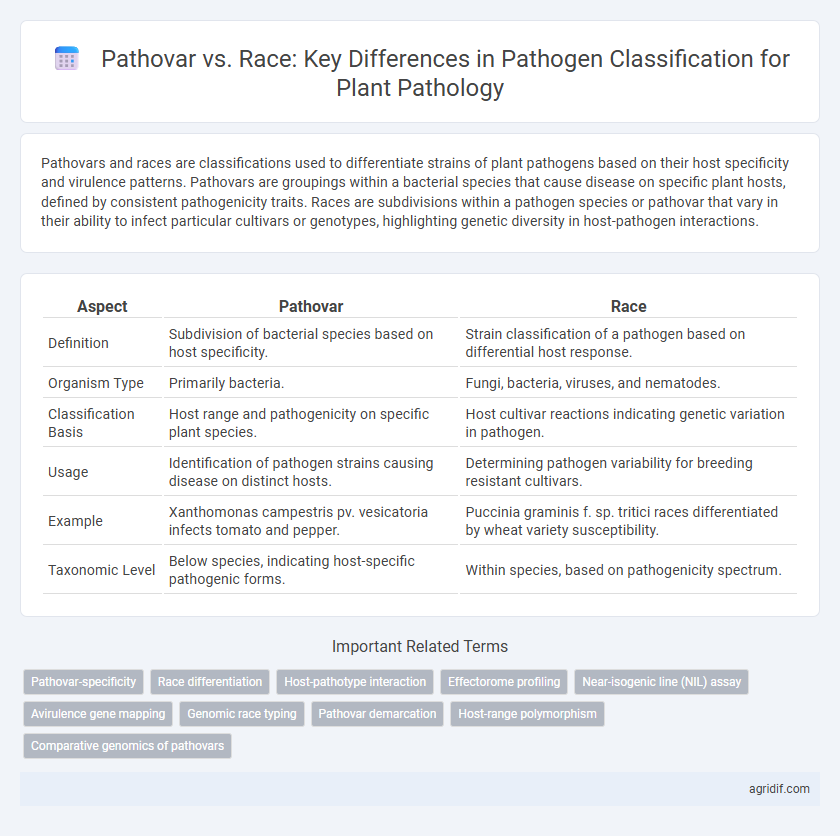
Pathovars and races are classifications used to differentiate strains of plant pathogens based on their host specificity and virulence patterns. Pathovars are groupings within a bacterial species that cause disease on specific plant hosts, defined by consistent pathogenicity traits. Races are subdivisions within a pathogen species or pathovar that vary in their ability to infect particular cultivars or genotypes, highlighting genetic diversity in host-pathogen interactions.
Table of Comparison
| Aspect | Pathovar | Race |
|---|---|---|
| Definition | Subdivision of bacterial species based on host specificity. | Strain classification of a pathogen based on differential host response. |
| Organism Type | Primarily bacteria. | Fungi, bacteria, viruses, and nematodes. |
| Classification Basis | Host range and pathogenicity on specific plant species. | Host cultivar reactions indicating genetic variation in pathogen. |
| Usage | Identification of pathogen strains causing disease on distinct hosts. | Determining pathogen variability for breeding resistant cultivars. |
| Example | Xanthomonas campestris pv. vesicatoria infects tomato and pepper. | Puccinia graminis f. sp. tritici races differentiated by wheat variety susceptibility. |
| Taxonomic Level | Below species, indicating host-specific pathogenic forms. | Within species, based on pathogenicity spectrum. |
Introduction to Pathogen Classification in Plant Pathology
Pathogen classification in plant pathology differentiates pathovars and races based on host specificity and genetic variations. Pathovars are bacterial strains distinguished by their ability to infect specific host species or varieties, while races are subsets within a pathogen species that exhibit differential virulence on distinct host genotypes. Understanding these distinctions enhances accurate diagnosis, management strategies, and breeding for disease resistance in crops.
Defining Pathovars: Concept and Significance
Pathovars are distinct bacterial strains differentiated by the specific plant hosts they infect, classified based on pathogenicity rather than genetic composition. This concept enables precise identification and management of pathogens affecting particular crops, improving disease control strategies. Defining pathovars is crucial for developing targeted resistance breeding and tailored agricultural practices to mitigate the impact of bacterial diseases.
Understanding Races: Definition and Role
Races in plant pathology refer to groups within a pathogen species differentiated by their specific ability to infect particular host plant varieties, often determined through host-pathogen interaction studies. Unlike pathovars, which classify pathogens based on the host plant species they infect, races highlight genetic variability and virulence differences within a pathogen population. Understanding races is crucial for breeding disease-resistant crops and developing targeted disease management strategies.
Pathovar vs Race: Key Differences
Pathovars are bacterial strains differentiated by their host specificity and pathogenicity on particular plant species, while races denote variations within a pathogen species based on differential virulence against host plant cultivars. Pathovar classification primarily relies on host range and symptom expression, whereas race classification is determined through host resistance gene interactions and differential pathogenicity tests. Understanding the distinction between pathovars and races is essential for effective disease management and breeding resistant crop varieties.
Criteria for Assigning Pathovar Status
Pathovar status is assigned based on host specificity and the ability of a pathogen to cause disease in distinct plant hosts, focusing on phenotypic characteristics and pathogenicity profiles. Pathovars are defined by differential interactions with specific plant species or cultivars, reflecting discrete genetic variations within a bacterial species. Unlike races, which are classified primarily based on virulence differences within the same host species, pathovars emphasize taxonomic distinctions tied to host range and host-pathogen interactions.
Criteria for Defining Races in Pathogens
Races in plant pathogens are defined based on their differential interactions with a set of host cultivars carrying specific resistance genes, characterized by unique virulence patterns. Pathovar classification relies on pathogenicity to particular host species or varieties, while race differentiation depends on the genetic specificity of the pathogen's ability to infect or overcome resistance within a single host species. Molecular markers and gene-for-gene relationships between host resistance genes and pathogen avirulence genes are critical criteria for distinguishing races in plant pathogen populations.
Genetic Basis of Pathovar and Race Differentiation
Pathovar and race classification in plant pathogens differ primarily in their genetic determinants and host specificity; pathovars are distinguished by genes controlling pathogenicity on specific plant species, while races differ based on genetic variation governing virulence on particular cultivars or resistance genes within a host species. Molecular markers such as avirulence (Avr) genes and effector proteins often define race variation by interacting with host resistance (R) genes, whereas pathovar differentiation involves broader genetic traits influencing host range and infection mechanisms. Understanding these genetic bases aids in developing targeted disease management strategies and breeding resistant crop varieties tailored to specific pathogen variants.
Implications for Disease Management
Pathovars classify pathogens based on host specificity, guiding targeted crop breeding for resistance, while races are differentiated by their ability to overcome specific resistance genes within a host species, informing rotation and deployment strategies. Understanding the distinction enhances precision in selecting resistant cultivars and designing integrated disease management plans. Effective control depends on continuously monitoring pathogen populations to detect emerging races and adapting strategies accordingly.
Case Studies: Pathovar and Race Classification in Major Crops
Pathovar classification in major crops like rice and tomato focuses on host specificity and pathogenicity patterns, differentiating strains based on the plant species they infect. Race classification, commonly used in wheat and corn, identifies pathogen groups based on their virulence against specific resistance genes in host plants. Case studies reveal that integrating both pathovar and race concepts enhances disease management strategies and breeding programs by targeting pathogen diversity at multiple levels.
Future Perspectives in Pathogen Classification
Emerging genomic technologies and machine learning algorithms are transforming pathogen classification by enabling precise identification of pathovars and races based on genetic markers and virulence factors. Integrating multi-omics data will refine the differentiation between closely related pathogen strains, enhancing disease management strategies. Future pathogen classification systems will prioritize dynamic, real-time surveillance to predict pathogen evolution and improve crop resistance breeding programs.
Related Important Terms
Pathovar-specificity
Pathovars classify bacterial pathogens based on host specificity and symptom expression, providing a precise framework for identifying strains that infect particular plant species. In contrast, races are defined by their interaction with specific host resistance genes, highlighting genetic variation in virulence within a pathogen species but not necessarily reflecting host range.
Race differentiation
Race differentiation in plant pathology classifies pathogen strains based on their specific virulence patterns against host plant resistance genes, enabling precise identification of pathogen variants within a species. Unlike pathovars, which are defined by host range, races are distinguished by their ability to overcome particular host resistance mechanisms, crucial for targeted disease management and breeding resistant crop varieties.
Host-pathotype interaction
Pathovars classify bacterial strains based on host range specificity determined by differential host-pathogen interactions, while races categorize fungal or oomycete pathogen isolates according to virulence profiles on specific host cultivars. The distinction hinges on host-pathotype interaction, with pathovars defined by their ability to infect distinct plant species and races identified through differential disease reactions in genetically defined host accessions.
Effectorome profiling
Effectorome profiling distinguishes pathovars by analyzing specific sets of secreted effectors that determine host range and virulence, whereas races are defined based on differential host resistance responses. This molecular approach enhances pathogen classification accuracy by linking effector repertoires to pathogenicity and host specificity within plant pathology.
Near-isogenic line (NIL) assay
Pathovars classify bacterial strains based on host specificity, while races distinguish fungal pathogen variants by differential host resistance; near-isogenic line (NIL) assays enable precise genetic dissection of host-pathogen interactions by comparing disease responses in host lines differing only at specific resistance loci. NIL assays are critical for identifying race-specific avirulence genes and validating pathovar-specific virulence factors in plant pathology.
Avirulence gene mapping
Pathovars are differentiated based on host range and pathogenicity traits, while races are classified by their avirulence gene profiles interacting with specific plant resistance genes. Avirulence gene mapping enables precise identification of race-specific effectors, facilitating targeted breeding for durable disease resistance in crops.
Genomic race typing
Pathovar classification categorizes bacterial pathogens based on host specificity and disease symptoms, while race classification identifies pathogen variants that differ in virulence on specific host cultivars. Genomic race typing utilizes whole-genome sequencing and comparative genomics to reveal genetic variations responsible for virulence differences, enabling precise identification of races at the molecular level for targeted disease management.
Pathovar demarcation
Pathovar classification in plant pathology is based on host specificity and distinct pathogenicity patterns on particular plant species, allowing precise identification of bacterial strains responsible for disease. Unlike race classification that primarily focuses on differential virulence within a single host species, pathovar demarcation emphasizes ecological and pathogenic distinctions across diverse host plants.
Host-range polymorphism
Pathovars represent bacterial strains distinguished by their distinct host range, reflecting host-range polymorphism at the species level, whereas races classify fungal or oomycete pathogens based on differential virulence patterns on specific host cultivars. Host-range polymorphism in pathovars underscores genetic adaptations enabling infection of diverse plant species, while races emphasize variation in pathogenicity within a single host species.
Comparative genomics of pathovars
Comparative genomics of pathovars reveals distinct genetic variations linked to host specificity and virulence factors, providing a more nuanced classification than traditional race-based systems. Pathovars are defined by characteristic pathogenicity patterns on specific host plants, while races reflect intra-pathovar variability, with genomic analyses enabling precise differentiation through effector gene profiles and genomic islands.
Pathovar vs Race for pathogen classification Infographic
 agridif.com
agridif.com