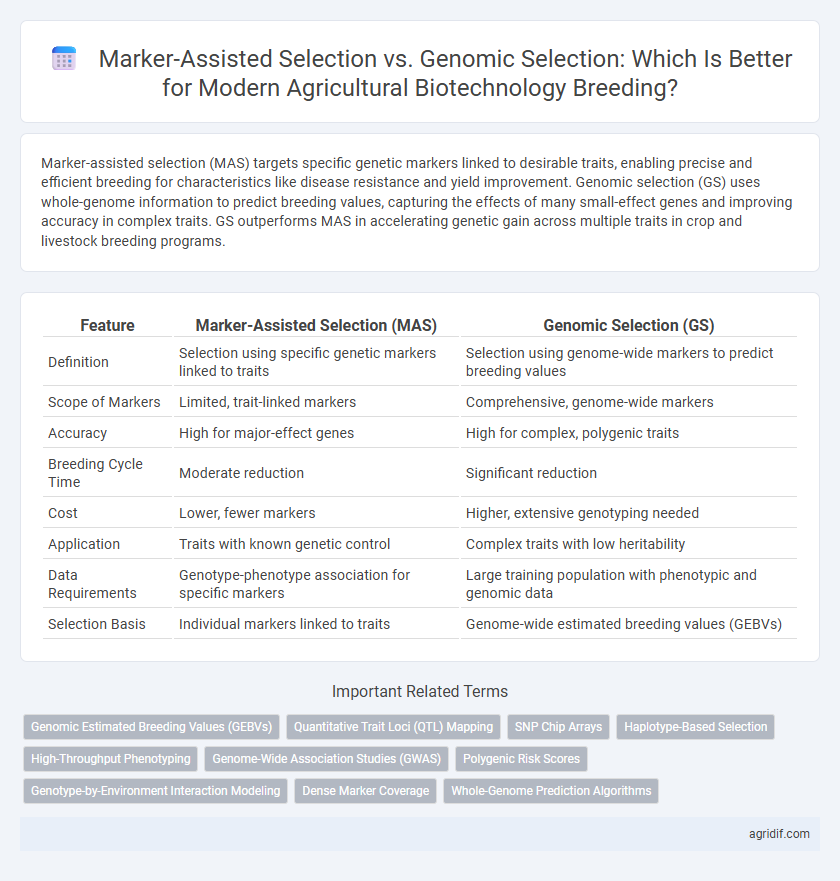
Marker-assisted selection (MAS) targets specific genetic markers linked to desirable traits, enabling precise and efficient breeding for characteristics like disease resistance and yield improvement. Genomic selection (GS) uses whole-genome information to predict breeding values, capturing the effects of many small-effect genes and improving accuracy in complex traits. GS outperforms MAS in accelerating genetic gain across multiple traits in crop and livestock breeding programs.
Table of Comparison
| Feature | Marker-Assisted Selection (MAS) | Genomic Selection (GS) |
|---|---|---|
| Definition | Selection using specific genetic markers linked to traits | Selection using genome-wide markers to predict breeding values |
| Scope of Markers | Limited, trait-linked markers | Comprehensive, genome-wide markers |
| Accuracy | High for major-effect genes | High for complex, polygenic traits |
| Breeding Cycle Time | Moderate reduction | Significant reduction |
| Cost | Lower, fewer markers | Higher, extensive genotyping needed |
| Application | Traits with known genetic control | Complex traits with low heritability |
| Data Requirements | Genotype-phenotype association for specific markers | Large training population with phenotypic and genomic data |
| Selection Basis | Individual markers linked to traits | Genome-wide estimated breeding values (GEBVs) |
Introduction to Marker-Assisted and Genomic Selection
Marker-Assisted Selection (MAS) utilizes specific DNA markers linked to desired traits to enhance breeding efficiency, enabling the targeted selection of individuals with favorable genetic profiles. Genomic Selection (GS) employs genome-wide marker data and advanced statistical models to predict breeding values, capturing the effects of numerous small-effect genes simultaneously. Both techniques accelerate genetic gain in crop and livestock breeding by increasing selection accuracy and reducing breeding cycle time compared to traditional phenotypic selection.
Principles of Marker-Assisted Selection in Plant Breeding
Marker-Assisted Selection (MAS) in plant breeding leverages molecular markers closely linked to desired traits to enhance selection accuracy and efficiency. By identifying specific DNA sequences associated with phenotypic traits, MAS enables early and precise selection of superior genotypes, reducing breeding cycle time. This approach significantly improves genetic gain by facilitating the simultaneous selection of multiple traits and minimizing environmental influence on phenotype expression.
Key Features of Genomic Selection in Agriculture
Genomic selection in agriculture utilizes genome-wide markers to predict the breeding value of plants, enabling faster and more accurate selection compared to Marker-Assisted Selection, which targets specific genes or QTLs. This approach leverages dense marker data and advanced statistical models, capturing minor and major gene effects simultaneously for complex traits like yield, disease resistance, and drought tolerance. High-throughput genotyping and phenotyping further enhance the precision and efficiency of genomic selection, accelerating genetic gain in crop improvement programs.
Comparing Technologies: MAS vs Genomic Selection
Marker-Assisted Selection (MAS) targets specific, known genetic markers linked to desirable traits, enabling precise selection but is limited to traits with identified markers. Genomic Selection (GS) utilizes genome-wide marker data to predict breeding values, capturing the effects of numerous small-effect genes and offering higher accuracy for complex traits. GS outperforms MAS in breeding efficiency by enabling earlier selection and accelerating genetic gain across diverse crop species.
Efficiency and Accuracy of Genetic Gain
Marker-Assisted Selection (MAS) targets specific genetic markers linked to desirable traits, enabling precise introgression but often captures limited genetic variance, which can constrain genetic gain efficiency. Genomic Selection (GS) utilizes genome-wide marker information to predict breeding values, significantly improving the accuracy of selection and accelerating genetic gain across complex traits. Empirical studies demonstrate GS outperforms MAS in both reliability and speed, especially for polygenic traits, making it a more efficient approach for modern crop and livestock breeding programs.
Cost-Effectiveness and Resource Requirements
Marker-Assisted Selection (MAS) offers cost-effectiveness by targeting a limited number of known genetic markers, reducing the need for extensive phenotyping and genotyping resources compared to Genomic Selection (GS). GS requires substantial investment in high-throughput genotyping and computational resources to analyze genome-wide markers but delivers higher accuracy in predicting breeding values for complex traits. The choice between MAS and GS depends on available budget, infrastructure, and breeding program goals, with MAS suitable for traits controlled by few genes and GS preferred for polygenic traits requiring comprehensive genomic information.
Applications in Crop and Livestock Improvement
Marker-Assisted Selection (MAS) enables precise identification of key genetic markers linked to desirable traits, accelerating the breeding of crops like maize and wheat with improved yield and disease resistance. Genomic Selection (GS) utilizes genome-wide marker data to predict breeding values, enhancing accuracy in developing livestock such as dairy cattle and pigs with superior growth and reproductive traits. Both MAS and GS are integral to modern agricultural biotechnology, driving sustainable crop productivity and livestock genetic gains through targeted breeding strategies.
Challenges and Limitations of Each Method
Marker-Assisted Selection (MAS) faces limitations such as reliance on a limited number of markers linked to target traits, which can miss complex polygenic traits and reduce accuracy in diverse genetic backgrounds. Genomic Selection (GS) requires extensive, high-quality genomic and phenotypic datasets for model training, leading to high costs and computational demands, and its prediction accuracy can decrease when applied across different populations or environments. Both methods grapple with challenges including environmental interactions and the integration of multi-trait selection, complicating their use in practical breeding programs.
Future Trends in Breeding Technologies
Future trends in breeding technologies emphasize the integration of Marker-Assisted Selection (MAS) and Genomic Selection (GS) to accelerate crop improvement with greater precision. Genomic Selection leverages high-density genome-wide markers to predict breeding values, outperforming MAS, which targets specific loci, especially for complex traits influenced by multiple genes. Advances in machine learning algorithms and high-throughput phenotyping are poised to enhance genomic prediction accuracy, enabling more efficient selection cycles and faster development of climate-resilient and high-yield crop varieties.
Conclusion: Integrating MAS and Genomic Selection
Integrating Marker-Assisted Selection (MAS) and Genomic Selection enhances crop breeding efficiency by leveraging the precise gene targeting of MAS and the comprehensive genome-wide predictions of genomic selection. Combining these methods accelerates the development of cultivars with improved yield, disease resistance, and environmental adaptability. This synergy supports sustainable agriculture by optimizing genetic gain and reducing breeding cycle time.
Related Important Terms
Genomic Estimated Breeding Values (GEBVs)
Genomic Estimated Breeding Values (GEBVs) leverage genome-wide marker information to predict the genetic potential of plants with higher accuracy than traditional Marker-Assisted Selection (MAS), enabling faster and more precise crop improvement. By integrating dense genotypic data and advanced statistical models, GEBVs facilitate the selection of complex traits controlled by multiple genes, enhancing the efficiency of breeding programs in agricultural biotechnology.
Quantitative Trait Loci (QTL) Mapping
Marker-Assisted Selection (MAS) utilizes Quantitative Trait Loci (QTL) mapping to identify specific genetic markers linked to desirable traits, enabling targeted breeding decisions and accelerating trait introgression. In contrast, Genomic Selection (GS) employs genome-wide marker data to predict breeding values without relying solely on identified QTLs, enhancing accuracy in selecting complex polygenic traits in crop improvement programs.
SNP Chip Arrays
Marker-Assisted Selection (MAS) utilizes a limited number of Single Nucleotide Polymorphism (SNP) markers identified through SNP chip arrays to track specific genes linked to desirable traits, making it efficient for traits controlled by a few major genes. In contrast, Genomic Selection (GS) leverages high-density SNP chip arrays to evaluate thousands of markers genome-wide, enabling more accurate prediction of complex traits influenced by multiple loci, thereby accelerating genetic gain in crop and livestock breeding programs.
Haplotype-Based Selection
Haplotype-based selection in agricultural biotechnology offers a refined approach compared to marker-assisted selection by capturing the combined effect of linked genetic variants across chromosome segments, improving the accuracy of predicting complex traits. Genomic selection leverages genome-wide marker data, but integrating haplotype information enhances predictive power for breeding by utilizing the structure of linkage disequilibrium and historical recombination events within populations.
High-Throughput Phenotyping
High-throughput phenotyping enhances marker-assisted selection by accelerating the identification of key genetic markers linked to desirable traits, while genomic selection integrates vast phenotypic datasets to improve prediction accuracy for complex traits in crop breeding. Leveraging advanced imaging and sensor technologies, both methods optimize genetic gain but genomic selection offers a more comprehensive approach by utilizing genome-wide marker data combined with precise phenotypic information.
Genome-Wide Association Studies (GWAS)
Marker-Assisted Selection (MAS) leverages identified single nucleotide polymorphisms (SNPs) linked to specific traits through Genome-Wide Association Studies (GWAS) to enhance breeding accuracy by targeting a limited set of significant markers, whereas Genomic Selection (GS) uses genome-wide SNP data from GWAS to predict breeding values of individuals, capturing the effects of both major and minor alleles and improving genetic gain in complex traits. GWAS provides critical high-density marker information enabling MAS to focus on key loci, while GS integrates entire genomic profiles for comprehensive selection models in crop improvement.
Polygenic Risk Scores
Marker-assisted selection (MAS) utilizes specific genetic markers linked to desirable traits, but genomic selection (GS) leverages genome-wide markers and polygenic risk scores (PRS) to predict breeding values more accurately for complex traits influenced by multiple genes. Polygenic risk scores, derived from dense SNP arrays, enhance the precision of GS by aggregating small effect sizes across the genome, enabling more effective selection in crop and livestock improvement programs.
Genotype-by-Environment Interaction Modeling
Genomic Selection integrates high-density molecular markers with phenotype and environment data to model genotype-by-environment interactions, enhancing prediction accuracy for complex traits across diverse conditions. Marker-Assisted Selection typically targets specific loci but lacks comprehensive GxE modeling, limiting its efficiency in breeding programs addressing variable environments.
Dense Marker Coverage
Dense marker coverage enhances the precision of genomic selection by capturing the cumulative effect of numerous small-effect genes across the entire genome, whereas marker-assisted selection relies on fewer, specific markers linked to major quantitative trait loci. This comprehensive genomic information in genomic selection enables higher prediction accuracy and accelerated breeding cycles, improving crop trait improvement efficiency.
Whole-Genome Prediction Algorithms
Whole-genome prediction algorithms in genomic selection utilize dense marker data across the entire genome to estimate breeding values with higher accuracy compared to marker-assisted selection, which relies on a limited number of markers linked to specific traits. These algorithms enable breeders to capture the effects of both major and minor genes, accelerating genetic gain and improving complex trait selection efficiency in crop and livestock breeding programs.
Marker-Assisted Selection vs Genomic Selection for Breeding Infographic
 agridif.com
agridif.com