Backcross breeding rapidly transfers specific disease resistance genes from a donor to an elite cultivar while preserving the latter's genetic background, making it ideal for introducing major resistance genes. Recurrent selection enhances polygenic resistance by gradually accumulating favorable alleles through repeated cycles of selection and intercrossing within a population, improving overall genetic diversity and durable resistance. Combining both methods can optimize resistance durability by integrating major gene effects with quantitative inheritance.
Table of Comparison
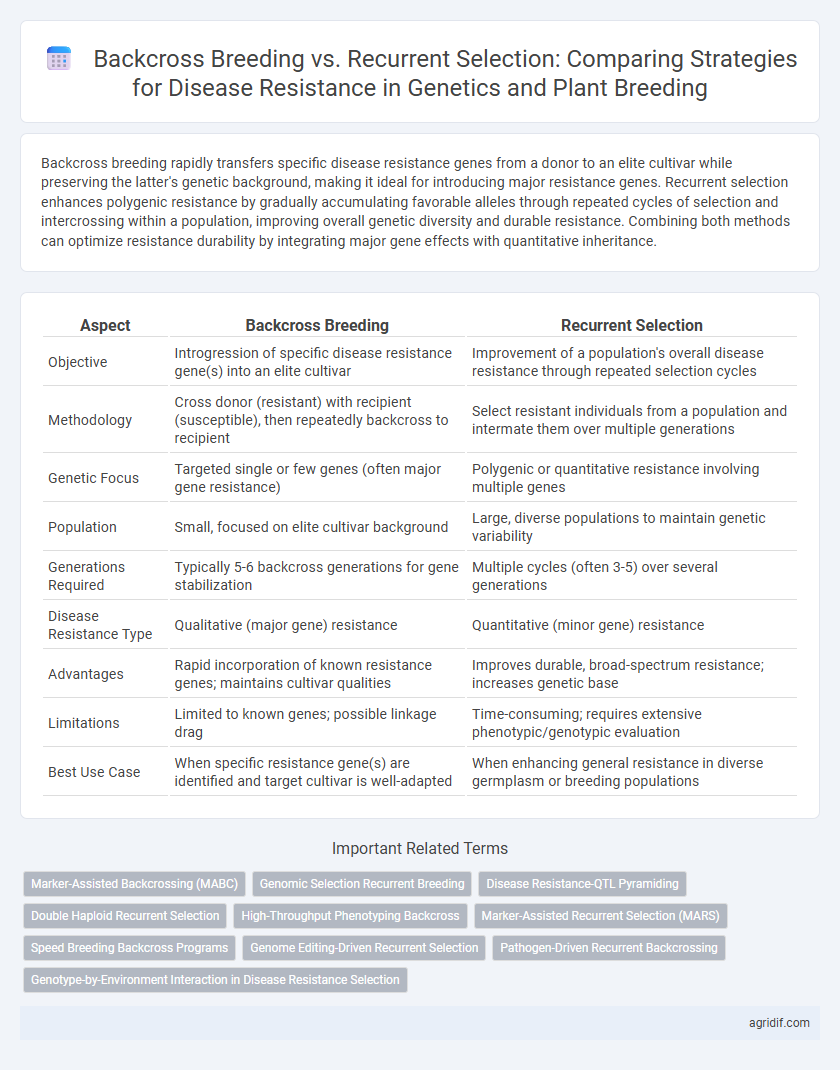
| Aspect | Backcross Breeding | Recurrent Selection |
|---|---|---|
| Objective | Introgression of specific disease resistance gene(s) into an elite cultivar | Improvement of a population's overall disease resistance through repeated selection cycles |
| Methodology | Cross donor (resistant) with recipient (susceptible), then repeatedly backcross to recipient | Select resistant individuals from a population and intermate them over multiple generations |
| Genetic Focus | Targeted single or few genes (often major gene resistance) | Polygenic or quantitative resistance involving multiple genes |
| Population | Small, focused on elite cultivar background | Large, diverse populations to maintain genetic variability |
| Generations Required | Typically 5-6 backcross generations for gene stabilization | Multiple cycles (often 3-5) over several generations |
| Disease Resistance Type | Qualitative (major gene) resistance | Quantitative (minor gene) resistance |
| Advantages | Rapid incorporation of known resistance genes; maintains cultivar qualities | Improves durable, broad-spectrum resistance; increases genetic base |
| Limitations | Limited to known genes; possible linkage drag | Time-consuming; requires extensive phenotypic/genotypic evaluation |
| Best Use Case | When specific resistance gene(s) are identified and target cultivar is well-adapted | When enhancing general resistance in diverse germplasm or breeding populations |
Introduction to Disease Resistance in Crop Improvement
Backcross breeding introduces specific disease resistance genes from a donor to an elite cultivar, enabling rapid incorporation of targeted resistance traits while maintaining the recipient genome's agronomic qualities. Recurrent selection enhances the frequency of multiple minor resistance alleles through repeated cycles of selection and recombination, promoting durable and broad-spectrum disease resistance in crop populations. Both methods are critical in modern crop improvement programs aiming to develop cultivars with effective and sustainable disease resistance.
Principles of Backcross Breeding in Plant Genetics
Backcross breeding in plant genetics involves repeatedly crossing a hybrid offspring with one of its parent plants to introduce or enhance specific disease resistance traits while retaining the parent's desirable characteristics. This method relies on precise selection of individuals carrying the target resistance allele, enabling the transfer of single genes or major genes with minimal disruption to the genetic background. Compared to recurrent selection, backcross breeding is more efficient for fixating disease resistance from donor lines into elite cultivars without extensive genetic variability.
Fundamentals of Recurrent Selection for Disease Resistance
Recurrent selection for disease resistance involves repeated cycles of selection and interbreeding to accumulate favorable alleles within a population, enhancing quantitative resistance traits. This method targets polygenic control of disease resistance, making it more effective for complex traits compared to backcross breeding, which introgresses specific major resistance genes from donor to recurrent parents. Recurrent selection improves overall genetic diversity and durability of resistance by continuously recombining resistant alleles, thereby reducing the risk of resistance breakdown under pathogen pressure.
Genetic Basis and Heritability of Disease Resistance Traits
Backcross breeding targets the introgression of specific disease resistance genes with high heritability into elite cultivars, ensuring rapid fixation of desired alleles through repeated hybridization with the recurrent parent. Recurrent selection exploits polygenic resistance governed by multiple genes with moderate to low heritability, enhancing overall population resistance via cyclical inter-mating and selection. The choice between these methods hinges on the genetic architecture of the resistance trait, with backcross breeding preferred for qualitative traits controlled by major genes and recurrent selection optimal for quantitative resistance involving complex gene interactions.
Efficiency of Backcross Breeding vs Recurrent Selection
Backcross breeding offers high efficiency for incorporating specific disease resistance genes into elite cultivars by repeatedly crossing progeny with the recurrent parent, ensuring rapid recovery of desired traits. In contrast, recurrent selection improves polygenic disease resistance through multiple cycles of selection and recombination, which is more time-consuming but enhances overall genetic diversity. The targeted gene transfer in backcross breeding leads to faster fixation of resistance traits, making it more efficient for monogenic disease resistance compared to the broader but slower gains achieved by recurrent selection.
Applicability to Different Disease Resistance Types
Backcross breeding is highly effective for incorporating major gene resistance traits into elite varieties, making it ideal for single-gene or qualitative disease resistance. Recurrent selection, on the other hand, enhances quantitative resistance controlled by multiple minor genes, improving polygenic disease resistance through successive cycles of selection. The choice between these methods depends on whether the targeted resistance is qualitative, favoring backcrossing for major genes, or quantitative, where recurrent selection builds durable resistance in crop populations.
Timeframe and Resources: Comparative Analysis
Backcross breeding rapidly introgresses specific disease resistance genes into elite cultivars within 3-5 generations, using targeted molecular markers that optimize resource allocation. Recurrent selection requires multiple cycles over 6-10 generations to accumulate polygenic resistance, demanding extensive field trials and phenotypic evaluations across diverse environments. Timeframe efficiency favors backcross breeding for single-gene resistance, while recurrent selection invests more resources for durable, quantitative disease resistance improvement.
Case Studies: Successful Implementation in Crops
Backcross breeding has been effectively utilized in rice to introduce blast disease resistance genes from wild relatives, resulting in cultivars with enhanced resistance and desirable agronomic traits. Recurrent selection demonstrated success in maize by progressively accumulating quantitative resistance alleles against gray leaf spot, improving population resistance over multiple cycles. These case studies highlight that backcross breeding excels in transferring specific major resistance genes, while recurrent selection is advantageous for enhancing polygenic disease resistance in crops.
Molecular Tools in Backcross and Recurrent Selection
Molecular tools in backcross breeding enable precise introgression of disease resistance genes by using marker-assisted selection (MAS) to track specific alleles, accelerating the recovery of recurrent parent genome while retaining resistance traits. In recurrent selection, molecular markers facilitate the identification and accumulation of favorable alleles across a population, enhancing polygenic disease resistance through genomic selection and genotyping-by-sequencing (GBS). The integration of high-throughput molecular markers and genome-wide association studies (GWAS) in both strategies improves accuracy, efficiency, and speed of breeding for durable disease resistance in crops.
Future Prospects and Challenges in Disease Resistance Breeding
Backcross breeding offers targeted introgression of specific disease resistance genes into elite cultivars, enabling precise improvement of resistance traits. Recurrent selection accelerates the accumulation of favorable alleles for polygenic disease resistance, enhancing overall population resilience. Future prospects include integrating genomic selection and CRISPR-based gene editing to overcome challenges like pathogen evolution and genetic bottlenecks in resistance breeding programs.
Related Important Terms
Marker-Assisted Backcrossing (MABC)
Marker-Assisted Backcrossing (MABC) accelerates the introgression of specific disease resistance genes into elite crop varieties by precisely tracking target alleles, offering higher efficiency compared to recurrent selection that relies on phenotypic evaluation over multiple cycles. While recurrent selection improves polygenic resistance through cumulative gene accumulation, MABC provides rapid incorporation and fixation of major resistance genes, making it a preferred strategy for targeted disease resistance breeding in plants.
Genomic Selection Recurrent Breeding
Genomic selection recurrent breeding enhances disease resistance in plants by predicting breeding values using genome-wide markers, accelerating the improvement process compared to traditional backcross breeding that introgresses specific resistance genes from donor to recurrent parents. This method increases genetic gain per cycle by capturing both major and minor effect loci, enabling more effective accumulation of polygenic resistance traits in breeding populations.
Disease Resistance-QTL Pyramiding
Backcross breeding efficiently introgresses specific disease resistance-QTLs into elite cultivars, ensuring targeted pyramiding of major resistance genes for enhanced durability. Recurrent selection accumulates multiple minor and moderate effect QTLs across populations, promoting polygenic resistance but requiring longer breeding cycles for effective disease resistance pyramiding.
Double Haploid Recurrent Selection
Backcross breeding effectively introgresses specific disease resistance genes into elite cultivars by rapidly recovering recurrent parent genome, while recurrent selection, particularly Double Haploid Recurrent Selection, accelerates genetic gain by combining multiple resistance alleles and fixing homozygosity through doubled haploids, enhancing selection efficiency and stability. Double Haploid Recurrent Selection outperforms traditional recurrent selection by producing uniform, homozygous lines in fewer generations, facilitating the development of durable disease-resistant plant varieties.
High-Throughput Phenotyping Backcross
Backcross breeding coupled with high-throughput phenotyping accelerates the introgression of specific disease resistance genes into elite plant varieties by enabling rapid, precise monitoring of phenotypic traits at large scales. In contrast, recurrent selection improves overall population resistance through multiple cycles of selection and recombination but is less efficient in targeting individual resistance loci compared to the precision of high-throughput phenotyping in backcross programs.
Marker-Assisted Recurrent Selection (MARS)
Backcross breeding efficiently introgresses specific disease resistance genes from donor to elite lines, while Marker-Assisted Recurrent Selection (MARS) combines genome-wide marker data to accumulate multiple minor resistance alleles across populations for durable resistance. MARS accelerates genetic gain by selecting individuals with favorable marker profiles linked to quantitative resistance loci, outperforming traditional phenotypic recurrent selection in complex traits like polygenic disease resistance.
Speed Breeding Backcross Programs
Backcross breeding accelerates disease resistance introgression by rapidly incorporating specific resistance genes from donor to elite lines, especially when combined with speed breeding techniques that shorten generation times. Recurrent selection improves polygenic resistance but typically requires multiple cycles and larger populations, making speed breeding backcross programs more efficient for targeting major resistance genes swiftly.
Genome Editing-Driven Recurrent Selection
Genome editing-driven recurrent selection accelerates the development of disease-resistant crop varieties by enabling precise modifications across multiple generations, enhancing allele frequency of resistance genes without introducing linkage drag common in backcross breeding. This method leverages CRISPR/Cas systems to systematically improve polygenic resistance traits, offering a more efficient and targeted alternative to traditional backcross breeding approaches.
Pathogen-Driven Recurrent Backcrossing
Pathogen-driven recurrent backcrossing enhances disease resistance by repeatedly introgressing specific resistance genes from donor to elite cultivars, ensuring targeted pathogen defense. This method contrasts with recurrent selection, which increases overall population resistance but lacks the precision and speed of allele fixation achieved through backcrossing in breeding programs.
Genotype-by-Environment Interaction in Disease Resistance Selection
Backcross breeding accelerates the introgression of specific disease resistance genes into elite genotypes, minimizing genotype-by-environment interaction effects by stabilizing resistance traits across diverse environments. Recurrent selection enhances population-wide resistance by exploiting genetic variation but requires multiple cycles to effectively address genotype-by-environment interactions, making it less efficient for rapidly fixing stable disease resistance.
Backcross breeding vs recurrent selection for disease resistance Infographic
 agridif.com
agridif.com