Backcrossing is a traditional method in genetics and plant breeding that introduces disease resistance genes from a donor parent into an elite cultivar while maintaining most of the cultivar's genome, ensuring the retention of desirable agronomic traits. Forward breeding accelerates the development of disease-resistant varieties by selecting progeny with favorable resistance genes from segregating populations, enhancing genetic diversity and adaptation. While backcrossing is precise and maintains cultivar integrity, forward breeding offers broader scope for combining multiple resistance genes from diverse sources.
Table of Comparison
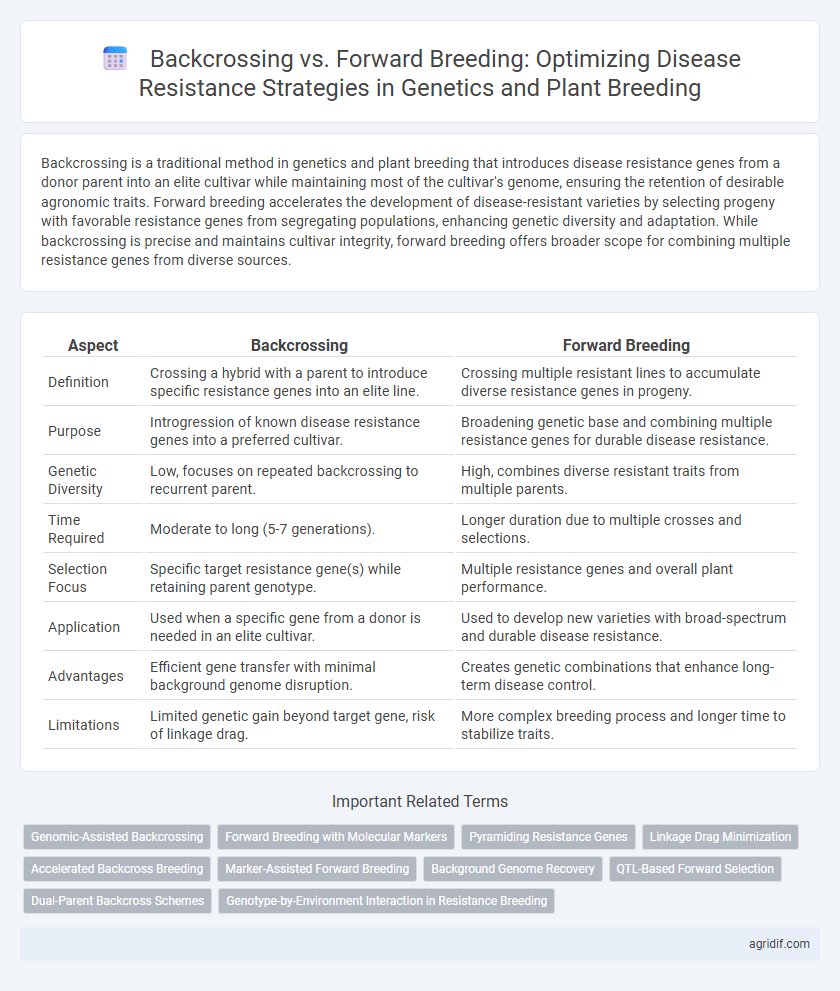
| Aspect | Backcrossing | Forward Breeding |
|---|---|---|
| Definition | Crossing a hybrid with a parent to introduce specific resistance genes into an elite line. | Crossing multiple resistant lines to accumulate diverse resistance genes in progeny. |
| Purpose | Introgression of known disease resistance genes into a preferred cultivar. | Broadening genetic base and combining multiple resistance genes for durable disease resistance. |
| Genetic Diversity | Low, focuses on repeated backcrossing to recurrent parent. | High, combines diverse resistant traits from multiple parents. |
| Time Required | Moderate to long (5-7 generations). | Longer duration due to multiple crosses and selections. |
| Selection Focus | Specific target resistance gene(s) while retaining parent genotype. | Multiple resistance genes and overall plant performance. |
| Application | Used when a specific gene from a donor is needed in an elite cultivar. | Used to develop new varieties with broad-spectrum and durable disease resistance. |
| Advantages | Efficient gene transfer with minimal background genome disruption. | Creates genetic combinations that enhance long-term disease control. |
| Limitations | Limited genetic gain beyond target gene, risk of linkage drag. | More complex breeding process and longer time to stabilize traits. |
Introduction to Disease Resistance in Plant Breeding
Disease resistance in plant breeding involves incorporating genetic traits that enable plants to withstand pathogen attacks, ensuring crop stability and yield. Backcrossing focuses on transferring specific resistance genes from a donor to a recurrent parent while maintaining elite agronomic traits, optimizing gene introgression efficiency. Forward breeding utilizes population improvement and selection to develop novel resistant varieties by combining multiple resistance alleles, enhancing long-term genetic diversity and durability against evolving pathogens.
Principles of Backcrossing in Agricultural Genetics
Backcrossing in agricultural genetics involves repeatedly crossing a hybrid offspring with one of its parents or a genetically similar individual to incorporate a specific disease resistance trait while retaining the recurrent parent's desirable characteristics. This method accelerates the transfer of resistance alleles by minimizing genetic variability outside the target gene region, facilitating precise introgression. The effectiveness of backcrossing depends on molecular markers to monitor genome recovery and ensure the desired gene is retained with minimal linkage drag.
Forward Breeding: Objectives and Methodologies
Forward breeding in plant genetics aims to incorporate novel disease resistance genes from diverse germplasm into elite cultivars by crossing and selecting progeny with enhanced resistance and desirable agronomic traits. Methodologies include marker-assisted selection (MAS) for early detection of resistance alleles, genomic selection to predict breeding values, and phenotypic screening under controlled pathogen exposure. This approach accelerates the development of varieties with durable resistance by pyramiding multiple genes and adapting to evolving pathogen populations.
Genetic Basis of Disease Resistance Traits
Backcrossing introduces a specific resistant gene from a donor parent into a recurrent parent genome, maintaining the overall genotype while enhancing disease resistance through targeted gene introgression. Forward breeding leverages recombination and selection among progeny with diverse genetic backgrounds, enabling the accumulation of multiple resistance alleles governed by quantitative trait loci (QTLs). Understanding the genetic architecture of resistance traits--whether monogenic or polygenic--determines the appropriate breeding strategy to achieve durable disease resistance.
Comparative Efficiency: Backcrossing vs Forward Breeding
Backcrossing efficiently transfers specific disease resistance genes from a donor to an elite cultivar, preserving the recipient's desirable genetic background while introducing targeted resistance traits. Forward breeding accelerates the accumulation of multiple resistance alleles through recurrent selection and recombination, promoting durable and broad-spectrum disease resistance. Backcrossing offers precision in gene introgression, whereas forward breeding enhances genetic diversity and adaptation to evolving pathogen pressures.
Marker-Assisted Selection in Backcrossing and Forward Breeding
Marker-assisted selection in backcrossing accelerates the introgression of specific disease resistance genes from donor to elite cultivars while minimizing linkage drag. In forward breeding, marker-assisted selection enables the simultaneous pyramiding of multiple resistance genes by tracking favorable alleles across segregating populations. Both approaches enhance precision in developing disease-resistant plant varieties, but backcrossing is preferred for incorporating known major resistance genes, whereas forward breeding suits complex, quantitative resistance traits.
Case Studies: Successes in Disease Resistance Breeding
Backcrossing has demonstrated significant success in disease resistance breeding, particularly in rice blast resistance where introgression of resistance genes like Pi9 from wild relatives improved cultivated varieties without compromising yield. Forward breeding in wheat has effectively combined multiple resistance genes against rust diseases by selecting progenies with complex gene interactions, thereby enhancing durable resistance. Case studies highlight that integrating backcrossing and forward breeding accelerates development of resistant cultivars by exploiting known resistance loci and novel gene combinations.
Genetic Diversity Outcomes in Backcrossing and Forward Breeding
Backcrossing typically results in reduced genetic diversity by repeatedly crossing a hybrid with one of its parents to introgress specific disease resistance genes, limiting the genetic variation in subsequent generations. Forward breeding maintains higher genetic diversity by selecting resistant individuals from diverse populations, enabling the accumulation of multiple resistance alleles and adaptive traits. The choice between backcrossing and forward breeding impacts long-term breeding program effectiveness and resilience against evolving plant pathogens.
Challenges and Limitations of Both Breeding Strategies
Backcrossing for disease resistance is limited by its dependence on a single resistant donor, leading to a narrowed genetic base and potential linkage drag that introduces undesirable traits. Forward breeding faces challenges such as longer breeding cycles and difficulty in pyramiding multiple resistance genes due to complex inheritance patterns. Both strategies require extensive phenotyping and marker-assisted selection to accurately identify and retain resistance loci, which can be resource-intensive and time-consuming.
Future Prospects in Disease Resistance Breeding Approaches
Backcrossing remains a valuable technique for introgressing specific disease resistance genes from donor to elite cultivars, ensuring trait stability across generations. Forward breeding accelerates the incorporation of multiple resistance genes through genomic selection and marker-assisted approaches, enhancing durability against evolving pathogens. Future prospects emphasize integrating high-throughput phenotyping and genome editing technologies to combine backcrossing precision with forward breeding efficiency for sustainable disease resistance.
Related Important Terms
Genomic-Assisted Backcrossing
Genomic-assisted backcrossing enhances traditional backcross methods by integrating molecular markers to accelerate the introgression of disease resistance genes into elite cultivars, ensuring precise gene transfer with minimal linkage drag. This approach outperforms forward breeding by reducing breeding cycles and increasing selection accuracy for resistance traits in complex genomes.
Forward Breeding with Molecular Markers
Forward breeding with molecular markers accelerates the development of disease-resistant plants by enabling precise selection of target genes during early generations, reducing the breeding cycle time. This approach enhances genetic gain by combining phenotypic selection with marker-assisted selection, improving the efficiency and accuracy of introducing resistance traits compared to traditional backcrossing methods.
Pyramiding Resistance Genes
Backcrossing efficiently introduces specific resistance genes from a donor parent into an elite cultivar, preserving most of the recurrent parent's genome, while forward breeding combines multiple resistance genes through successive hybridizations to develop plants with broad-spectrum disease resistance. Pyramiding resistance genes via forward breeding enhances durability against diverse pathogens by stacking multiple resistance loci, whereas backcrossing is ideal for quickly incorporating known, single resistance traits.
Linkage Drag Minimization
Backcrossing effectively incorporates disease resistance genes from donor plants into elite cultivars while minimizing linkage drag by repeatedly crossing progeny with the recurrent parent, thereby preserving desirable agronomic traits. Forward breeding, however, often retains larger donor genome segments around resistance loci, increasing the risk of linkage drag and potentially introducing unwanted traits alongside disease resistance.
Accelerated Backcross Breeding
Accelerated backcross breeding enhances disease resistance by rapidly introgressing specific resistance genes from a donor to a recipient plant while preserving the recipient's elite genetic background, significantly reducing breeding cycles compared to traditional backcrossing. Forward breeding, although effective for developing new gene combinations, requires more time and resources, making accelerated backcrossing a preferred strategy for targeted introgression with precise genetic control in crop improvement.
Marker-Assisted Forward Breeding
Marker-assisted forward breeding accelerates the development of disease-resistant plant varieties by enabling precise selection of multiple resistance genes simultaneously, enhancing genetic gain and durability. Unlike backcrossing, which primarily introgresses single resistance genes into elite lines, forward breeding combines novel resistance alleles from diverse germplasm using molecular markers, reducing linkage drag and increasing breeding efficiency.
Background Genome Recovery
Backcrossing enhances disease resistance by recurrently crossing offspring with the resistant parent, prioritizing background genome recovery up to 90-99% within 5-6 generations. Forward breeding accelerates the introgression of resistance traits but often retains a larger proportion of donor genome, resulting in less efficient background genome recovery compared to backcrossing.
QTL-Based Forward Selection
QTL-based forward selection in plant breeding enables precise introgression of disease resistance genes by continuously selecting progeny with favorable quantitative trait loci, accelerating the development of resistant cultivars compared to traditional backcrossing methods. This approach enhances genetic gain by integrating multi-gene resistance and maintaining a broader genetic base, improving durability and adaptability against evolving pathogens.
Dual-Parent Backcross Schemes
Dual-parent backcross schemes enhance disease resistance by integrating favorable alleles from both parents into elite cultivars through repeated backcrossing cycles, ensuring the retention of desired agronomic traits while introducing specific resistance genes. This approach contrasts with forward breeding, which typically involves recurrent selection in progeny populations but may not maintain the elite genetic background as effectively, making dual-parent backcrossing superior for targeted gene introgression in crop improvement programs.
Genotype-by-Environment Interaction in Resistance Breeding
Backcrossing efficiently introgresses specific resistance genes into elite cultivars, but its success is often limited by genotype-by-environment interactions that affect the stability of disease resistance across diverse conditions. Forward breeding, by selecting within segregating populations under multiple environmental conditions, enhances the durability of resistance by capturing favorable allelic combinations responsive to genotype-by-environment variation.
Backcrossing vs Forward Breeding for Disease Resistance Infographic
 agridif.com
agridif.com