Qualitative traits in genetics and plant breeding are controlled by one or a few genes, resulting in distinct categories with clear phenotypic differences. Quantitative traits, however, are governed by multiple genes and environmental factors, producing continuous variation across a range. Understanding the distinction between these trait types is crucial for effective breeding strategies and accurate genetic analysis.
Table of Comparison
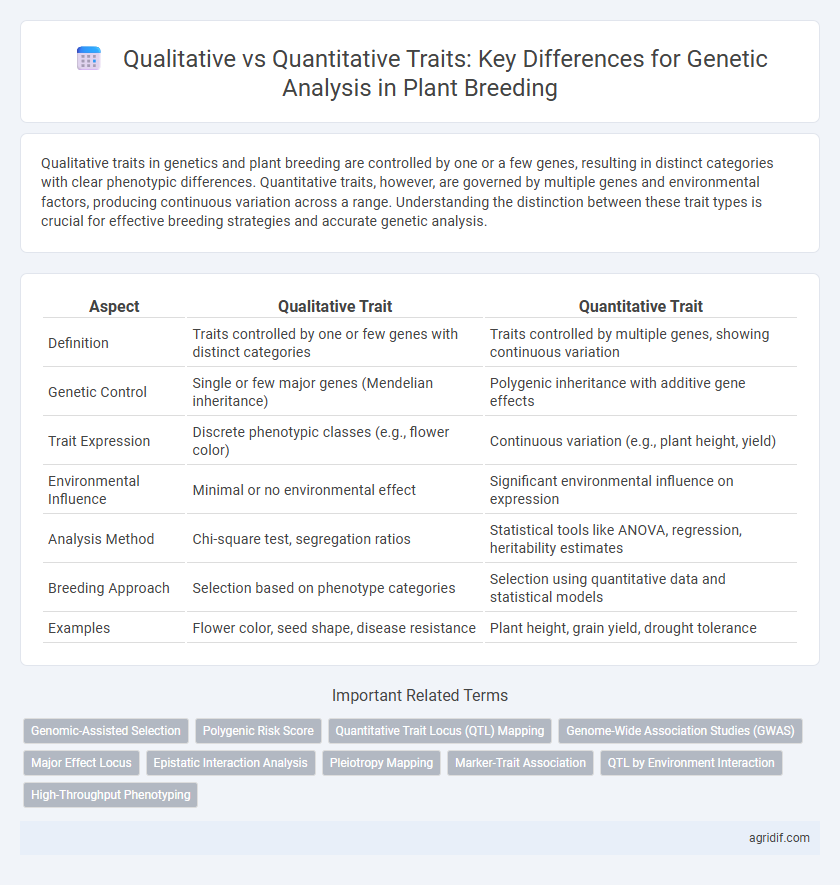
| Aspect | Qualitative Trait | Quantitative Trait |
|---|---|---|
| Definition | Traits controlled by one or few genes with distinct categories | Traits controlled by multiple genes, showing continuous variation |
| Genetic Control | Single or few major genes (Mendelian inheritance) | Polygenic inheritance with additive gene effects |
| Trait Expression | Discrete phenotypic classes (e.g., flower color) | Continuous variation (e.g., plant height, yield) |
| Environmental Influence | Minimal or no environmental effect | Significant environmental influence on expression |
| Analysis Method | Chi-square test, segregation ratios | Statistical tools like ANOVA, regression, heritability estimates |
| Breeding Approach | Selection based on phenotype categories | Selection using quantitative data and statistical models |
| Examples | Flower color, seed shape, disease resistance | Plant height, grain yield, drought tolerance |
Introduction to Qualitative and Quantitative Traits
Qualitative traits in genetics and plant breeding are controlled by one or a few genes, exhibiting distinct phenotypic categories such as flower color or seed shape. Quantitative traits arise from the combined effect of multiple genes and environmental factors, resulting in continuous variation like plant height or yield. Understanding the genetic basis of these traits is essential for effective selection and breeding strategies aimed at crop improvement.
Genetic Basis of Qualitative Traits
Qualitative traits in genetics and plant breeding are controlled by one or a few genes with distinct alleles, resulting in discrete phenotypic categories such as flower color or seed shape. These traits exhibit clear Mendelian inheritance patterns, making them easier to analyze through classical genetic methods. The simplicity of their genetic basis allows breeders to predict trait segregation and perform marker-assisted selection effectively.
Genetic Basis of Quantitative Traits
Quantitative traits are controlled by multiple genes, each contributing small effects, resulting in continuous variation in phenotypes such as plant height or yield. These polygenic traits are often influenced by environmental factors, complicating genetic analysis compared to qualitative traits governed by single genes with discrete phenotypes. Understanding the genetic basis of quantitative traits involves mapping quantitative trait loci (QTL) to identify genomic regions associated with phenotypic variation in plant breeding.
Inheritance Patterns: Mendelian vs Polygenic
Qualitative traits in genetics and plant breeding follow Mendelian inheritance patterns, characterized by discrete phenotypic categories controlled by single genes with clear dominant and recessive alleles. Quantitative traits exhibit polygenic inheritance, governed by multiple genes with additive effects, resulting in continuous variation influenced by environmental factors. Understanding these inheritance modes is essential for effective selection and breeding strategies targeting specific traits.
Methods for Analyzing Qualitative Traits
Qualitative traits in genetics and plant breeding are typically analyzed using Mendelian segregation ratios and chi-square tests to determine the inheritance patterns of discrete phenotypic categories. Marker-assisted selection and linkage analysis are effective methods for identifying genes or loci associated with these qualitative traits, facilitating precise genetic mapping. Controlled crosses and segregation analysis remain foundational techniques for confirming dominant, recessive, or epistatic relationships among qualitative trait alleles.
Approaches for Studying Quantitative Traits
Quantitative traits in genetics and plant breeding are controlled by multiple genes and exhibit continuous variation, making their analysis complex compared to qualitative traits, which are governed by single genes with discrete phenotypes. Approaches for studying quantitative traits include QTL mapping, genome-wide association studies (GWAS), and genomic selection, all aimed at identifying genetic loci contributing to phenotypic variability. Statistical methods such as mixed linear models and interval mapping are essential for dissecting the genetic architecture of quantitative traits and improving breeding programs.
Phenotypic Variation and Its Measurement
Qualitative traits exhibit distinct phenotypic categories controlled by one or few genes, resulting in discrete variation, while quantitative traits show continuous phenotypic variation influenced by multiple genes and environmental factors. Phenotypic variation in qualitative traits is measured through categorical classification, whereas quantitative traits require statistical tools like variance analysis and heritability estimates to quantify trait distribution. Understanding these differences is critical for accurate genetic analysis and effective selection in plant breeding programs.
Molecular Markers in Trait Analysis
Qualitative traits are controlled by a few genes with major effects, while quantitative traits result from the combined influence of many genes, each contributing small effects. Molecular markers such as SSRs, SNPs, and AFLPs enable precise identification and mapping of genes associated with both qualitative and quantitative traits, facilitating marker-assisted selection. These markers enhance the efficiency of genetic analysis by linking phenotypic variation to specific genomic regions, accelerating the breeding of desired traits.
Implications for Plant Breeding Programs
Qualitative traits in plant breeding, controlled by one or a few genes with distinct phenotypic classes, facilitate straightforward selection processes due to their simple inheritance patterns. Quantitative traits, influenced by multiple genes and environmental factors, require statistical tools like QTL mapping and genomic selection to capture genetic variation effectively for complex traits such as yield and stress tolerance. Understanding the genetic architecture of these traits informs breeding strategies, enabling targeted improvement of crop performance and adaptation through marker-assisted or genomic-assisted selection.
Challenges in Genetic Analysis of Qualitative and Quantitative Traits
Genetic analysis of qualitative traits is complicated by Mendelian inheritance patterns but is often limited by environmental influence masking phenotypic distinctions. Quantitative traits involve multiple gene loci with additive effects, making it difficult to identify specific genes responsible due to gene-gene interactions and environmental variability. Statistical models such as QTL mapping and genome-wide association studies are required to address the complexity in dissecting quantitative trait loci under polygenic control.
Related Important Terms
Genomic-Assisted Selection
Qualitative traits in genetic analysis exhibit discrete variation controlled by a few genes, simplifying marker-assisted selection, whereas quantitative traits involve continuous variation influenced by multiple genes and environmental factors, necessitating genomic-assisted selection methods like genomic selection for accurate prediction. Genomic-assisted selection leverages high-density molecular markers across the genome to estimate breeding values for complex quantitative traits, significantly accelerating genetic gains in plant breeding programs.
Polygenic Risk Score
Qualitative traits are controlled by single genes with distinct phenotypic categories, while quantitative traits result from the combined effect of multiple genes and environmental factors, making them polygenic. Polygenic risk scores aggregate the effects of numerous genetic variants to predict the likelihood of complex traits, providing a powerful tool for genetic analysis and plant breeding strategies.
Quantitative Trait Locus (QTL) Mapping
Quantitative Trait Locus (QTL) mapping identifies specific genomic regions associated with variation in quantitative traits, such as yield or height, which are controlled by multiple genes and environmental factors. This approach utilizes molecular markers and statistical methods to dissect the genetic architecture of complex traits, enhancing precision in plant breeding programs.
Genome-Wide Association Studies (GWAS)
Qualitative traits, controlled by single or few genes with distinct phenotypic classes, contrast with quantitative traits influenced by multiple genes exhibiting continuous variation, posing unique challenges for Genome-Wide Association Studies (GWAS) in plant breeding. GWAS effectively maps quantitative trait loci (QTLs) by correlating genetic markers with phenotypic variation across diverse populations, enabling precise identification of genes underlying complex traits essential for crop improvement.
Major Effect Locus
Qualitative traits are controlled by one or a few major effect loci with clear Mendelian inheritance patterns, making them simpler to analyze genetically. Quantitative traits result from the combined influence of multiple minor effect loci, exhibiting continuous variation and complex polygenic inheritance.
Epistatic Interaction Analysis
Epistatic interaction analysis reveals the complex genetic architecture underlying quantitative traits, where multiple genes interact non-additively to influence phenotypic variation, unlike qualitative traits controlled by single genes with discrete effects. Understanding these gene-gene interactions enhances the precision of marker-assisted selection and genomic prediction in plant breeding for traits such as yield, drought tolerance, and disease resistance.
Pleiotropy Mapping
Qualitative traits are controlled by a few genes with major effects, making pleiotropy mapping more straightforward by linking single loci to multiple phenotypic traits, while quantitative traits involve polygenic inheritance with complex interactions that require advanced statistical models to dissect pleiotropic gene effects accurately. Understanding the genetic architecture of pleiotropy in quantitative traits aids in identifying key genomic regions influencing multiple agronomic characteristics, enhancing precision in plant breeding programs.
Marker-Trait Association
Qualitative traits, controlled by single genes, show clear marker-trait associations enabling straightforward genetic analysis, whereas quantitative traits involve multiple genes with small effects, requiring statistical models like QTL mapping to detect marker-trait associations. High-density markers and genome-wide association studies (GWAS) improve resolution in identifying loci linked to complex quantitative traits in plant breeding programs.
QTL by Environment Interaction
Quantitative Trait Loci (QTL) by Environment interaction significantly influences the expression of quantitative traits in plants, revealing how genetic factors vary across different environmental conditions. Understanding this interaction is crucial for genetic analysis and plant breeding, as it helps identify stable QTLs and develop cultivars with enhanced adaptability and performance under diverse environmental stresses.
High-Throughput Phenotyping
High-throughput phenotyping enables precise measurement of quantitative traits, such as yield and stress tolerance, that are controlled by multiple genes and exhibit continuous variation. In contrast, qualitative traits, determined by single genes with discrete phenotypic categories, require less complex phenotyping but benefit from molecular marker-assisted selection in genetic analysis.
Qualitative Trait vs Quantitative Trait for Genetic Analysis Infographic
 agridif.com
agridif.com