Quantitative Trait Loci (QTL) mapping identifies genomic regions linked to specific traits by analyzing controlled crosses, providing high power to detect loci with large effects. Genome-wide Association Studies (GWAS) scan natural populations to find marker-trait associations across the entire genome, capturing the genetic architecture of complex traits with higher resolution. Combining QTL mapping and GWAS enhances the accuracy and efficiency of trait dissection, accelerating genetic improvement in plant breeding programs.
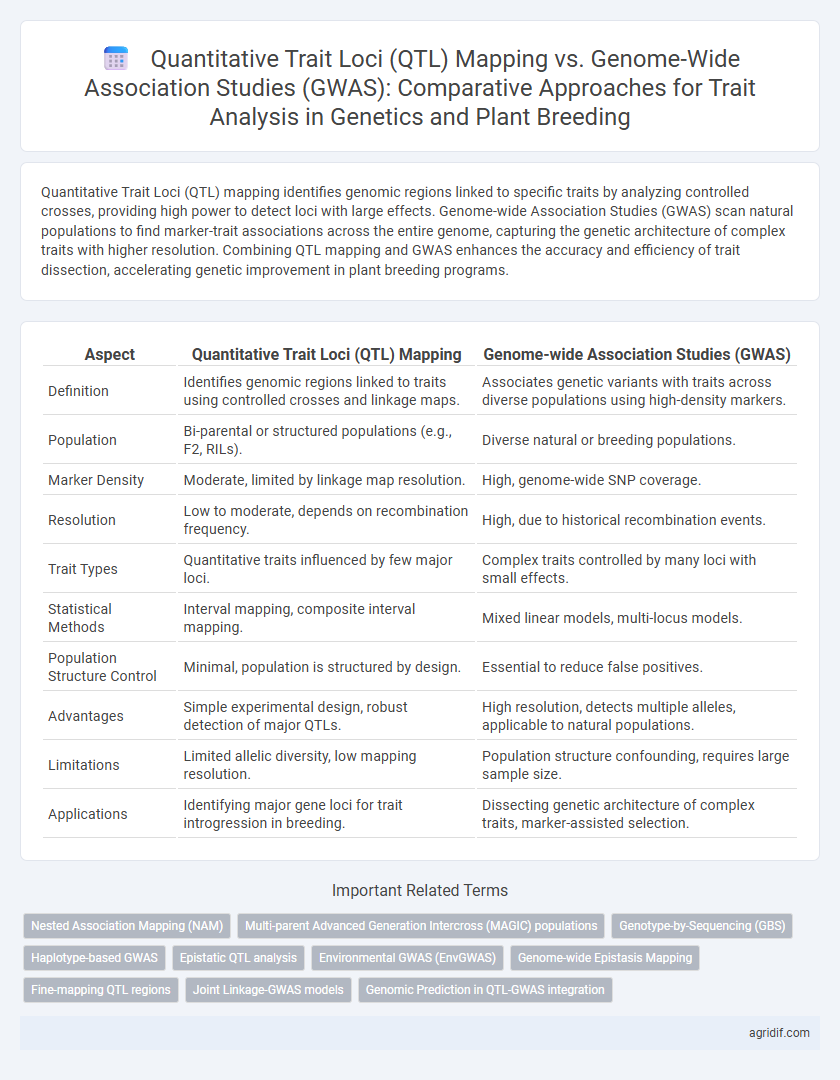
Table of Comparison
| Aspect | Quantitative Trait Loci (QTL) Mapping | Genome-wide Association Studies (GWAS) |
|---|---|---|
| Definition | Identifies genomic regions linked to traits using controlled crosses and linkage maps. | Associates genetic variants with traits across diverse populations using high-density markers. |
| Population | Bi-parental or structured populations (e.g., F2, RILs). | Diverse natural or breeding populations. |
| Marker Density | Moderate, limited by linkage map resolution. | High, genome-wide SNP coverage. |
| Resolution | Low to moderate, depends on recombination frequency. | High, due to historical recombination events. |
| Trait Types | Quantitative traits influenced by few major loci. | Complex traits controlled by many loci with small effects. |
| Statistical Methods | Interval mapping, composite interval mapping. | Mixed linear models, multi-locus models. |
| Population Structure Control | Minimal, population is structured by design. | Essential to reduce false positives. |
| Advantages | Simple experimental design, robust detection of major QTLs. | High resolution, detects multiple alleles, applicable to natural populations. |
| Limitations | Limited allelic diversity, low mapping resolution. | Population structure confounding, requires large sample size. |
| Applications | Identifying major gene loci for trait introgression in breeding. | Dissecting genetic architecture of complex traits, marker-assisted selection. |
Introduction to Trait Analysis in Plant Breeding
Quantitative Trait Loci (QTL) mapping and Genome-wide Association Studies (GWAS) are key methods for dissecting complex traits in plant breeding by identifying genetic variants associated with phenotypic variation. QTL mapping uses bi-parental populations to detect linkage between markers and traits, offering high power but limited resolution, while GWAS analyzes diverse natural populations to achieve higher resolution by exploiting historical recombination events. Combining QTL mapping and GWAS enhances the accuracy of trait analysis, accelerating marker-assisted selection and genomic prediction for crop improvement.
Overview of Quantitative Trait Loci (QTL) Mapping
Quantitative Trait Loci (QTL) mapping identifies chromosomal regions associated with phenotypic variation in traits by analyzing genetic markers in controlled breeding populations. This technique relies on linkage analysis to detect statistical associations between markers and quantitative traits, providing insights into the genetic architecture of complex traits. High-resolution QTL mapping enables the detection of loci influencing yield, disease resistance, and stress tolerance in crops, facilitating marker-assisted selection in plant breeding programs.
Fundamentals of Genome-wide Association Studies (GWAS)
Genome-wide Association Studies (GWAS) leverage high-density genetic markers to identify associations between genetic variants and phenotypic traits across diverse populations. Unlike Quantitative Trait Loci (QTL) mapping, which relies on biparental crosses, GWAS utilizes natural variation and linkage disequilibrium, enabling higher resolution mapping of complex traits. This approach facilitates the detection of multiple loci with small effects, advancing marker-assisted selection and genomic prediction in plant breeding programs.
Methodological Differences Between QTL Mapping and GWAS
QTL mapping relies on linkage analysis within biparental populations, identifying genomic regions associated with traits through recombination events, whereas GWAS utilizes natural populations and exploits historical recombination to detect marker-trait associations at higher resolution. QTL mapping typically has lower resolution but greater power to detect rare alleles, while GWAS offers fine mapping and genome-wide coverage but can be confounded by population structure and requires larger sample sizes. Both methods complement each other by balancing limitations in allele frequency detection, mapping precision, and population diversity in trait analysis.
Strengths and Limitations of QTL Mapping
QTL Mapping excels in identifying genomic regions associated with traits in controlled, bi-parental populations, providing high power to detect loci with large effects and facilitating the study of gene-by-environment interactions. However, its resolution is limited due to low recombination events, resulting in broad confidence intervals that hinder precise gene identification. The method also struggles with detecting loci of small effect and requires extensive phenotyping across multiple generations, making it less scalable compared to GWAS.
Advantages and Challenges of GWAS
Genome-wide Association Studies (GWAS) enable high-resolution mapping of complex traits by analyzing natural genetic variation across diverse populations, facilitating the identification of multiple loci associated with phenotypic traits. The main advantage of GWAS lies in its ability to detect common variants with small effects without requiring prior knowledge of the trait's genetic architecture. Challenges include the need for large sample sizes to achieve statistical power, the confounding effects of population structure, and the difficulty in pinpointing causal variants due to linkage disequilibrium.
Comparative Power and Resolution: QTL Mapping vs GWAS
Quantitative Trait Loci (QTL) mapping offers high statistical power for detecting loci with large effects by analyzing controlled crosses, but it often provides lower resolution due to limited recombination events and smaller population sizes. Genome-wide Association Studies (GWAS) leverage natural populations with extensive historical recombination, achieving higher mapping resolution and enabling detection of loci with small effects across the genome. While QTL mapping excels in identifying major effect alleles within biparental populations, GWAS offers increased resolution and power to capture diverse allelic variation in complex traits across diverse germplasm.
Applications in Crop Improvement and Variety Development
Quantitative Trait Loci (QTL) mapping employs controlled cross populations to identify genomic regions linked to complex traits, essential for marker-assisted selection in crop improvement. Genome-wide Association Studies (GWAS) utilize natural population diversity and high-density markers to detect trait-associated loci with higher resolution, accelerating variety development by pinpointing candidate genes. Both approaches complement each other in integrating genetic information for enhancing yield, stress tolerance, and quality traits in breeding programs.
Integrating QTL Mapping and GWAS for Enhanced Trait Dissection
Integrating Quantitative Trait Loci (QTL) mapping with Genome-wide Association Studies (GWAS) enhances the resolution and power of identifying genetic variants linked to complex traits in plants. QTL mapping leverages controlled crosses to detect loci with large effects, while GWAS analyzes natural populations to capture numerous small-effect variants and population diversity. Combining these approaches facilitates precise trait dissection, accelerates marker-assisted selection, and improves the efficiency of breeding programs targeting quantitative traits in crop species.
Future Perspectives in Genetic Trait Analysis
Future perspectives in genetic trait analysis emphasize integrating Quantitative Trait Loci (QTL) mapping and Genome-wide Association Studies (GWAS) to enhance precision breeding in plants. Advances in high-throughput sequencing and phenotyping technologies enable more accurate identification of complex trait loci, facilitating the discovery of novel alleles with agronomic importance. Machine learning algorithms and multi-omics data integration are poised to revolutionize predictive modeling of trait heritability and environmental interactions, accelerating crop improvement efforts.
Related Important Terms
Nested Association Mapping (NAM)
Nested Association Mapping (NAM) combines the strengths of Quantitative Trait Loci (QTL) mapping and Genome-wide Association Studies (GWAS) by utilizing structured populations derived from multiple parental lines to enhance the resolution and power of trait dissection. This approach overcomes limitations in linkage disequilibrium and genetic diversity, enabling precise identification of loci controlling complex traits in crops such as maize and wheat.
Multi-parent Advanced Generation Intercross (MAGIC) populations
Multi-parent Advanced Generation Intercross (MAGIC) populations combine the advantages of Quantitative Trait Loci (QTL) mapping and Genome-wide Association Studies (GWAS) by providing high-resolution genetic recombination and allele diversity, enhancing trait dissection accuracy. These populations enable more precise identification of complex trait loci through increased genetic variation and controlled population structure, outperforming traditional bi-parental QTL mapping and conventional GWAS in plant breeding programs.
Genotype-by-Sequencing (GBS)
Genotype-by-Sequencing (GBS) enhances both Quantitative Trait Loci (QTL) mapping and Genome-wide Association Studies (GWAS) by providing high-density, cost-effective single nucleotide polymorphism (SNP) markers critical for precise trait dissection in plant breeding. While QTL mapping leverages GBS data in controlled biparental populations to identify loci with large effects, GWAS utilizes the extensive genetic diversity captured through GBS in natural populations to detect numerous small-effect loci associated with complex traits.
Haplotype-based GWAS
Haplotype-based GWAS enhances trait analysis by examining combinations of alleles across multiple loci, improving the resolution and power to detect quantitative trait loci (QTL) linked to complex traits in plant genomes. This approach leverages linkage disequilibrium patterns to identify candidate genes more precisely than traditional single-marker QTL mapping, facilitating advancement in genetics and plant breeding.
Epistatic QTL analysis
Epistatic QTL analysis in quantitative trait loci mapping identifies interactions between multiple genomic regions influencing complex traits, enhancing the understanding of non-additive genetic effects in plant breeding. Genome-wide association studies (GWAS) offer high-resolution detection of epistatic interactions by leveraging natural population diversity and dense marker coverage, facilitating precise trait dissection and improved marker-assisted selection.
Environmental GWAS (EnvGWAS)
Quantitative Trait Loci (QTL) mapping identifies genomic regions linked to phenotypic variation in controlled crosses, while Genome-wide Association Studies (GWAS) analyze natural populations to detect marker-trait associations across the genome. Environmental GWAS (EnvGWAS) extends GWAS by incorporating environmental variables, enabling the dissection of genotype-environment interactions critical for understanding adaptive traits in diverse ecosystems.
Genome-wide Epistasis Mapping
Genome-wide epistasis mapping identifies interactions between multiple loci influencing quantitative traits by scanning the entire genome to detect non-additive effects missed by traditional QTL mapping. Combining GWAS with epistasis analysis enhances the understanding of complex genetic architectures controlling plant traits, enabling more precise marker-assisted selection in breeding programs.
Fine-mapping QTL regions
Fine-mapping QTL regions enhances the resolution of genetic loci associated with complex traits by integrating high-density markers and recombination events, enabling precise identification of candidate genes. GWAS complements this process by leveraging natural population diversity and linkage disequilibrium patterns to detect trait-associated variants at a genome-wide scale, facilitating the validation and refinement of QTL hotspots.
Joint Linkage-GWAS models
Joint Linkage-GWAS models integrate high-resolution marker data and linkage information to enhance the power and precision of detecting Quantitative Trait Loci (QTL) in diverse plant populations. These models leverage the advantages of traditional QTL mapping and genome-wide association studies (GWAS), enabling the identification of complex trait-associated loci with improved accuracy for plant breeding applications.
Genomic Prediction in QTL-GWAS integration
Integrating Quantitative Trait Loci (QTL) mapping with Genome-wide Association Studies (GWAS) enhances genomic prediction accuracy by combining the high-resolution marker-trait associations from GWAS with the linkage information from QTL mapping. This synergistic approach allows for more precise identification of causal variants and improves the efficiency of marker-assisted selection in plant breeding programs.
Quantitative Trait Loci (QTL) Mapping vs Genome-wide Association Studies (GWAS) for Trait Analysis Infographic
 agridif.com
agridif.com